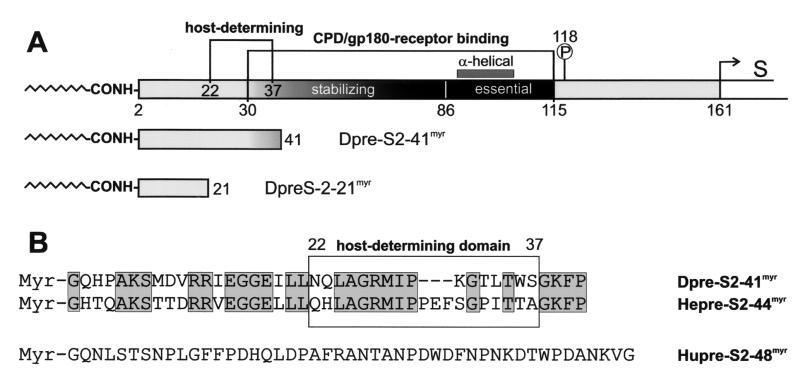
FIG. 1.
Schematic illustration of the pre-S domain of the DHBV L protein and the two DHBV pre-S-derived myristoylated peptides Dpre-S2-41myr and Dpre-S2-21myr (A) and the primary sequences of myristoylated Dpre-S2-41myr, Hepre-S2-44myr, and Hupre-S2-48myr (B) used in DHBV infection competition experiments. (A) The duCPD (gp180) receptor binding site (amino acids 30 to 115) within the pre-S domain (amino acids 1 to 161) of the DHBV L protein consists of an essential, partially α-helical subdomain (amino acids 86 to 115) and a most-randomly structured element (amino acids 30 to 85) which is needed for the formation of a high-affinity complex. Directly adjacent to the receptor binding site is Ser-118, which becomes phosphorylated (13). Partially overlapping with the stabilizing element but clearly extending it at its N terminus is the sequence that recovers infectivity of HHBV for PDHs (amino acids 22 to 37). (B) Sequence alignment of the two inhibitory peptides Dpre-S2-41myr and the homologous Hepre-S2-44myr. Sequence identity is 57%. Note that within the host-determining region (boxed sequence between amino acids 22 and 37), HHBV pre-S contains the additional insertion 31-PEF-33. The HBV pre-S-derived peptide Hupre-S2-48myr displays no significant homology to its avian counterparts.