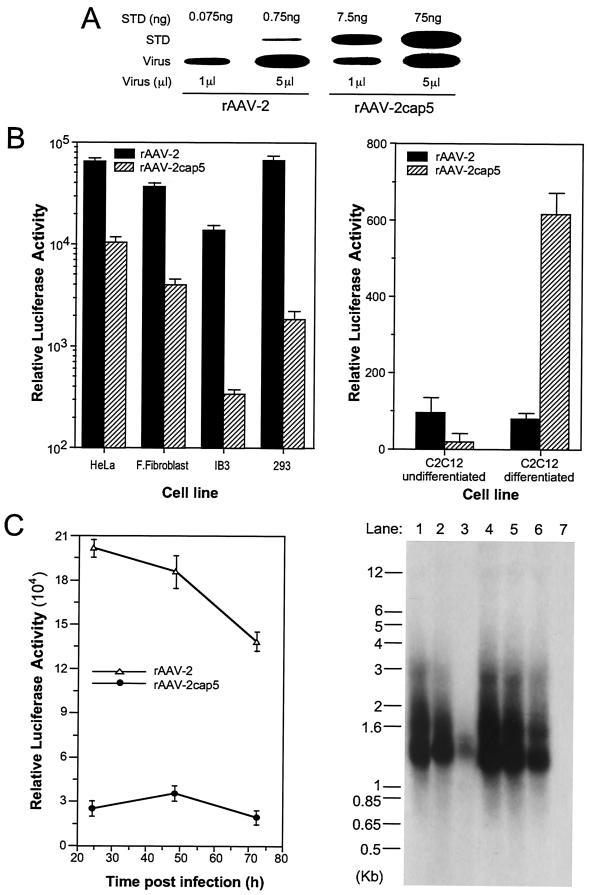
FIG. 2.
Evaluation of rAAV-2 and pseudotyped rAAV-2cap5 vectors. Both rAAV-2 and pseudotyped AAV-5 (rAAV-2cap5) contained the same luciferase reporter gene derived from the proviral plasmid pcisAV2RSVluc. The titers of both viral stocks were adjusted to equivalent physical particles per milliliter. (A) Titration of these two recombinant viral stocks by slot blot analysis against plasmid DNA standards (STD). The quantity of plasmid and the volume of virus evaluated in each well are marked above and below the blot, respectively. (B) Differences in transduction efficiencies following infection with either rAAV-2 or pseudotyped rAAV-2cap5 in a series of cell types (HeLa cells, primary fetal fibroblasts, IB3 cells, 293 cells, and undifferentiated or differentiated C2C12 muscle cells). Experiments were performed by infecting cells with 5 × 108 total particles/well in 12-well plates. Luciferase activity (relative light units/well) was determined 24 h after infection. Data represent the mean and standard error of the mean for four independent experiments. (C) Time course of transgene expression and viral genome persistence in HeLa cells following infection with rAAV-2 or rAAV-2cap5. A total of 109 particles of rAAV-2 or rAAV-2cap5 were used for infection of six-well plates. (Left panel) Luciferase activity was assayed at 24, 48, and 72 h postinfection. Data represent the mean and standard error of the mean for three independent experiments. (Right panel) Low-molecular-weight Hirt DNA was harvested from infected HeLa cells at 24 h (lanes 1 and 4), 48 h (lanes 2 and 5), and 72 h (lanes 3 and 6) and separated on a 1% agarose gel for Southern blotting and hybridization with a 32P-labeled luciferase probe. Each lane consists of Hirt DNA from one-half of a 35-mm dish of infected cells (note that microgram normalization of the DNA load did not provide an accurate comparison because cells were actively dividing over the time course of the study and contaminant genomic DNA in the Hirt DNA is dependent on cell number). Lanes 1 to 3, rAAV-2-infected cells; lanes 4 to 6, rAAV-2cap5-infected cells; 7, noninfected negative control.