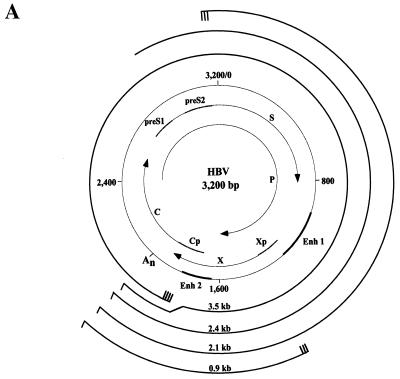
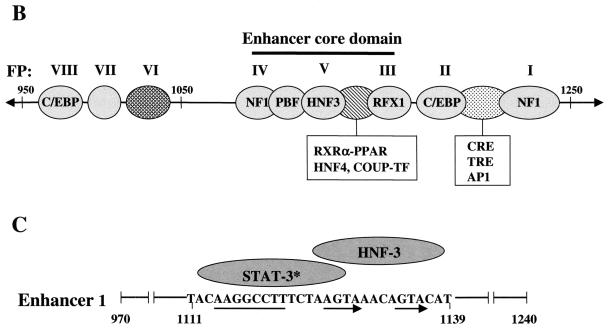
FIG. 1.
Schematic representations of the HBV genome and enhancer 1 element. (A) The viral genome is numbered (0 to 3,200 bp) according to the adw2 subtype of HBV. S, C, P, and X represent the viral genes encoding the surface antigen, core or e antigen, DNA polymerase, and X proteins, respectively. The HBV promoters (preS1, preS2, Cp, and Xp) and enhancers (Enh 1 and Enh 2) are indicated. An represents the single polyadenylation site used by all of the HBV RNAs. Each viral transcript is labeled with its respective length in kilobases (kb) and a poly(A) tail at the 3′ end. Multiple transcription initiation sites are indicated at the 5′ end of the 3.5-, 2.1-, and 0.9-kb RNAs. (B) Protein binding sites on Enh 1 are shown and footprint designations previously defined (38) are indicated above the corresponding sites. (C) The core domain spans nt 1080 to 1165 and is composed of the STAT-3 binding site, which is located at nt 1111 to 1127. The HNF-3 binding site spans nt 1124 to 1139. The DNA sequence of the STAT-3/HNF-3 region is shown with the corresponding cellular factors as indicated above. The thin line and arrows below the STAT-3-HNF-3 region indicate the positions of STAT-3 and FPV direct repeats, respectively.