Fig. 10.
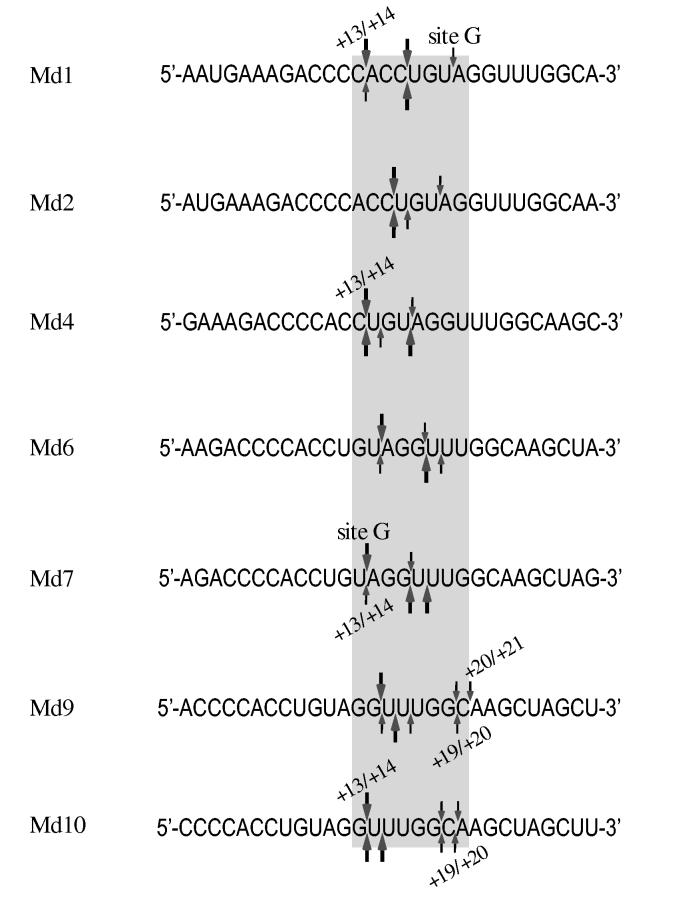
Comparison of HIV-1 and M-MuLV RNase H 5′ end-directed cleavages in the sequences of RNAs Md1 - Md10. The sequences of the 29-mer RNAs Md1 through Md10 are aligned by the RNA 5′ ends to compare the positions of 5′ end-directed cleavage sites. In each sequence, the extent of cleavage at a site is indicated as strong (large arrows) or medium (small arrows) for HIV-1 reverse transcriptase (above) or M-MuLV reverse transcriptase (below). As described in the Discussion, the range of the closest and furthest independent 5′ end-directed cleavage sites is indicated by the positions of the bordering nucleotides from the RNA 5′ end, the position of site G in substrates Md1 and Md7 is indicated, and the grey box highlights nucleotide positions +13 and +20 that include the range of distances where the 5′ end-directed cleavages occur.
