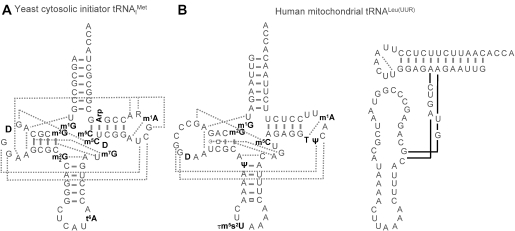
Figure 3.
Slight deviations from the classical tRNA structure. (A) Secondary structure of initiator from yeast. Modified nucleotides are in bold. Tertiary interactions are indicated by dotted lines. Note the absence of T54 and Ψ55 and the additional tertiary interactions between the A20 and the T-loop as compared to tRNAPhe. R denotes a purine at position 59 of for which sequence data in the literature are contradictory (96,89). (B) Human mitochondrial tRNALeu(UUR). The secondary structure of the native tRNA including all modified bases is shown on the left. Modified nucleotides are in bold and tertiary interactions are indicated by dotted lines. The loose structure of the anticodon domain of the unmodified transcript (98) is shown on the right. Modified nucleotides are abbreviated according to references (96,122).