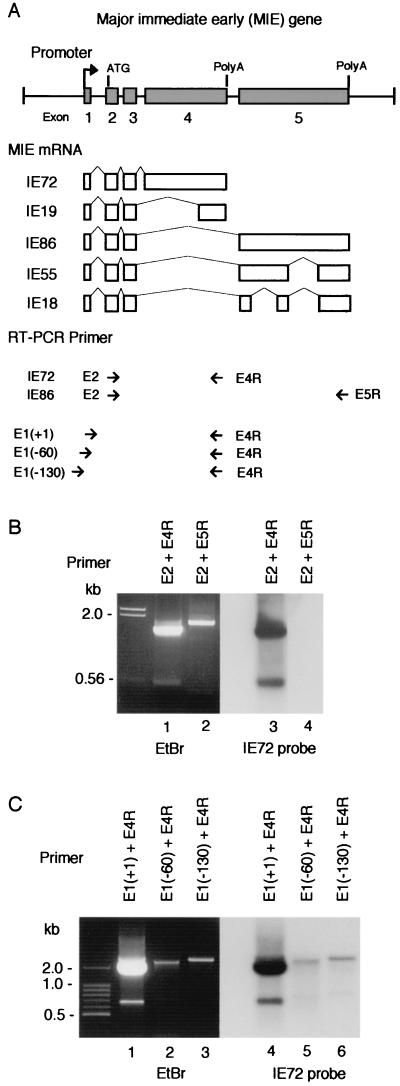
FIG. 1.
RT-PCR analysis of MIE mRNA. (A) MIE gene and the primers used for RT-PCR analysis. (Top) Major exon-intron structure of the MIE gene. Numbered solid boxes indicate each exon. The initiation codon (ATG) is located in exon 2. Both exon 4 and exon 5 have a stop codon and a poly(A) additional signal. (Middle) Alternative splicing to produce MIE mRNAs. (Bottom) Short arrows show position and direction of RT-PCR primers. The name of each primer is indicated. E1(+1) primer covers the transcription initiation site, and E1(−60) and E1(−130) primers cover to nt −60 and to nt −130 in the 5′-flanking region of the initiation site, respectively. (B and C) Results of RT-PCR analysis. RT-PCR was performed with total RNA (1 μg) of HCMV-infected HEL cells prepared at 48 hpi and with the primers indicated in each lane. RT-PCR products were examined by agarose gel electrophoresis (ethidium bromide [EtBr]) and Southern blot hybridization with an IE72-specific probe (IE72 probe). The positions of nucleotide markers are indicated. The nucleotide sequences of these RT-PCR primers and probes are shown in Table 1.