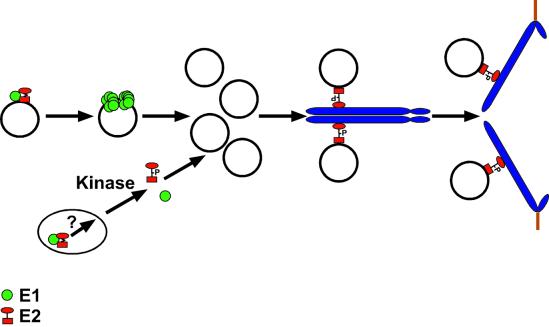
FIG. 8.
Model for the function of the E1 and E2 proteins in viral replication and nuclear retention of the viral genome during mitosis. E1 (green circles) and E2 (red circle/square) interaction facilitates the origin-specific binding of E1 to the viral DNA. It is believed that E2 must be removed in order for E1 to form double hexamers and perform its unwinding functions on viral origins for replication of the viral genome to occur (19). We propose that a second pool of E1-E2 complexes is present in the nucleus, especially when E1 levels are higher, as in the mutants. An unknown pathway that includes phosphorylation modification of the hinge region of E2 helps to disassemble such complexes. E2 can then bind to the pool of viral DNA that accumulates after S phase and, through interaction of the transactivation domain with a chromosomal receptor, associate with mitotic chromosomes. Phosphorylation of E2 proteins peaks in G2/M phase (15a). However, we do not know if chromosomally bound E2 is indeed phosphorylated. As the cellular chromosomes separate in mitosis, the tethered viral DNA is also segregated, ensuring that each daughter cell contains a nuclear copy of the viral genome following cell division.