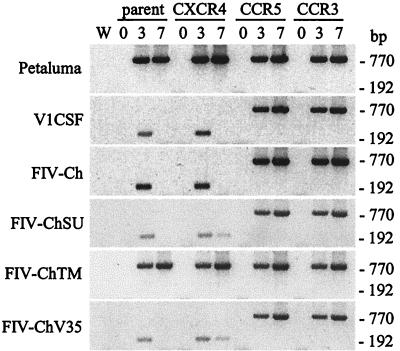
FIG. 7.
Detection of FIV sequences in trCrFK cells. Cells were infected with 104.5 (V1CSF, Petaluma, and FIV-Ch) or 102.5 (FIV-ChSU, -ChTM, and -ChV35) TCID50 of FIV/ml, and the FIV pol gene was detected in genomic DNA by single-round (770 bp) or nested (192 bp) PCR at 3 and 7 days p.i. trCrFK cell lines used included nontransfected parent cells and cells transfected with human CCR3, CXCR4, or CCR5. Amplification of a water blank (W) ensured the lack of contamination. FIV sequences were detected by a single round of PCR amplification in all Petaluma- and FIV-ChTM-infected cell lines and in cells expressing either CCR3 or CCR5 that were infected with the other viruses. In contrast, nested PCR was required to detect FIV DNA in parent cells and in cells expressing human CXCR4 following infection with V1CSF, FIV-Ch, FIV-ChSU, or FIV-ChV35.