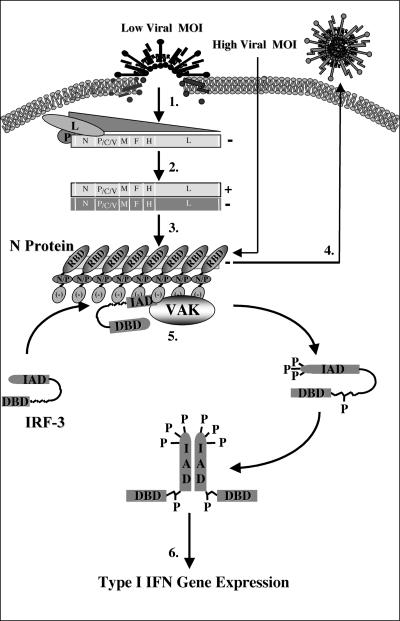
FIG. 8.
Schematic representation of IRF-3 activation following MeV infection. Following viral binding and fusion, the genome of MeV is released into the cytoplasm in tight association with the nucleocapsid (N) protein. Upon entry, N dissociates from the negative-stranded template as it is transcribed by the packaged polymerase composed of both the large (L) and phospho- (P) proteins of MeV (step 1). Transcription induces a gradient of protein production transcribing the highest amounts of the 3′ N gene and the lowest amount of the 5′-most L gene. As intracellular N protein concentrations rise, N associates with the genome template, causing a switch from transcription to replication by inducing the production of readthrough (+) full-length genome templates (step 2). Positive RNA synthesis serves as template for the production of full-length negative genomes, which, upon synthesis, are tightly bound by the N protein (step 3). Newly synthesized negative-strand genomes, associated with N, bind additional viral proteins in cooperation with numerous cellular proteins, allowing progeny virus to bud from the infected cell (step 4). During the course of infection, IRF-3 physically associates with the N protein of MeV through the interferon association domain (IAD) and its C-terminal negatively charged domain, leading to the C-terminal phosphorylation of IRF-3 by the virus-activated kinase (VAK) (step 5). Phosphorylation of IRF-3 is followed by its release from N, IRF dimerization, nuclear translocation, DNA binding, association with CBP, and transcriptional activation of the IFN-α/β genes (step 6). IAD, IRF association domain; DBD, DNA binding domain; RBD, RNA binding domain; N/P, hydrophobic domain involved in N/P interactions; and [−], negatively charged domain.