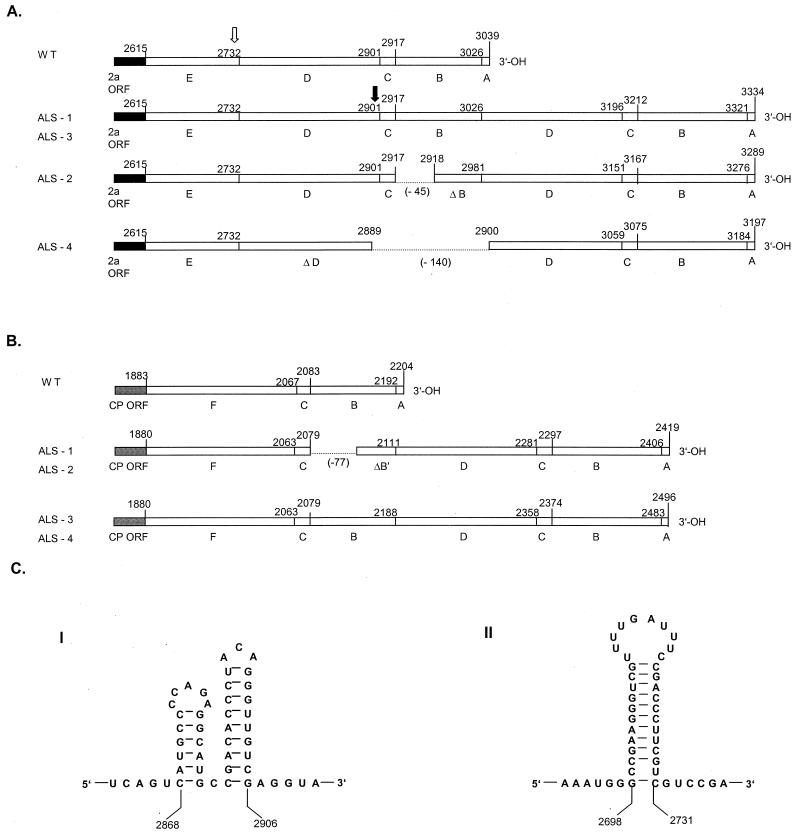
FIG. 2.
Schematic representations of the 3′ NTR of RNA 2 (A) and RNA 3 (B) of alstroemeria-infecting CMV strains and predicted secondary structures between nt 2868 and 2906 (I) and nt 2698 and 2731 (II) of wild-type (WT) RNA 2 (C). (A and B) Black and gray boxes indicate parts of the ORFs of 2a and CP genes, respectively. The numbers above the empty frames are the nucleotide numbers of the various regions relative to the 5′ terminus. The letters under the diagrams indicate the arbitrary regions of various lengths: A, 13 nt; B, 109 nt; C, 16 nt; D, 170 nt; E, 116 nt; and F, 183 nt. Regions with the same letter are identical in nucleotide sequence and length. The dotted lines indicate deletions (lengths are in parentheses). Δ indicates partially deleted regions. The open arrowhead indicates stem loop structure II (shown in panel C), where replication of RNA 2 is proposed to halt prior to restarting downstream of that position to form a recombinant RNA 2. The solid arrowhead indicates the position of stem loop structure I (C), where replication termination is thought to occur prior to reinitiation on RNA 3.