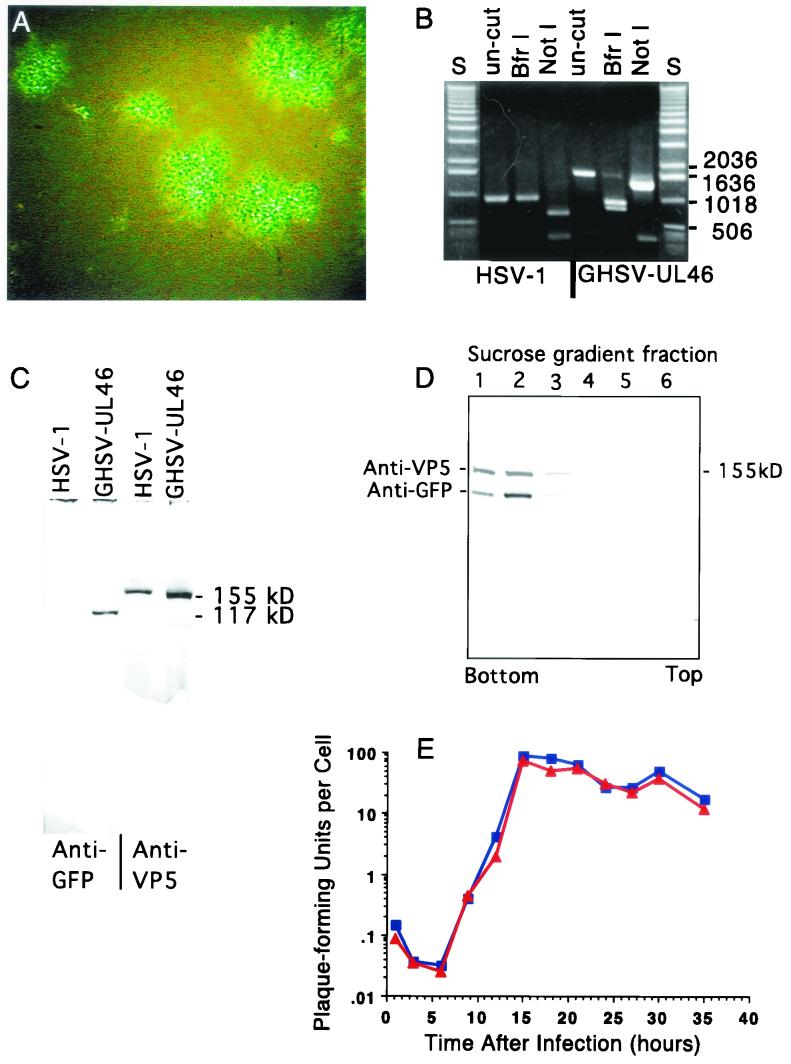
FIG.2.
Characteristics of GHSV-UL46. (A) Green fluorescent plaques generated by GHSV-UL46 viewed with a fluorescence microscope 4 days after a monolayer of Vero cells was infected with dilute GHSV-UL46. (B) Ethidium bromide-stained agarose gel showing a segment of DNA amplified from wild-type HSV-1 or GHSV-UL46 DNA with primers 9 and 10 (lanes un-cut) and restriction digests of the amplified band cut with BfrI or NotI. The primers for the PCR were complementary to sequences on each side of the targeted site of insertion of GFP (Fig. 1). S, standards of the indicated lengths (base pairs). (C) Western blot showing that partially purified preparations of GHSV-UL46 contain a polypeptide (electrophoretic mobility corresponding to an apparent molecular mass of 110 to 120 kDa) reactive with an antibody specific for GFP that is absent in wild-type HSV-1 (left two lanes). Both preparations contain a polypeptide that reacts with an antibody against the major capsid protein VP5 (right two lanes). (D) Density gradient equilibrium sedimentation illustrating similar buoyant-density distributions of the VP11/12-GFP fusion protein and VP5, the major capsid protein that served as a marker for virions. VP11/12-GFP and VP5 were identified by the staining of a Western blot of the sucrose gradient fractions with specific antibodies against GFP and VP5. The discontinuous gradient was formed from steps (from the bottom) of 77, 50, 40, 30, and 20% (wt/vol) sucrose. (E) One-step growth curve comparing growth of wild-type HSV-1 to that of GHSV-UL46. Subconfluent Vero cells were infected at a multiplicity of 7 PFU/cell. At 3-h intervals, total PFU were measured.