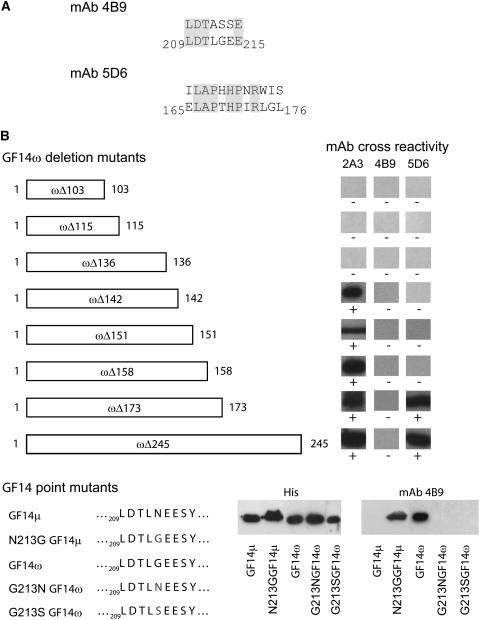
Figure 3.
GBC mAbs epitopes map to three distinct discontinuous sequences within the Arabidopsis 14-3-3s. A, GBC mAbs were used to screen phage display heptamer and dodecamer peptide libraries. Screening of 1 × 1011 clones identified peptides with a consensus sequence corresponding to Arabidopsis GF14 sequences for two of the antibodies. B, Truncations of GF14ω were used to localize the 2A3 epitope and confirm the phage display identification of the 4B9 and 5D6 epitopes. PCR was used to generate C-terminal truncation constructs of GF14ω, which were examined by western analysis. C, Mutation of GF14μ and GF14ω were used to further confirm and establish a key residue for the 4B9 GBC mAb epitope. Strand overlap extension PCR was used with Arabidopsis GF14μ to produce an Asn to Gly mutation and GF14ω to produce a Gly to Ser or Asn mutation at residue 213 in the constructs, respectively.