Figure 3.
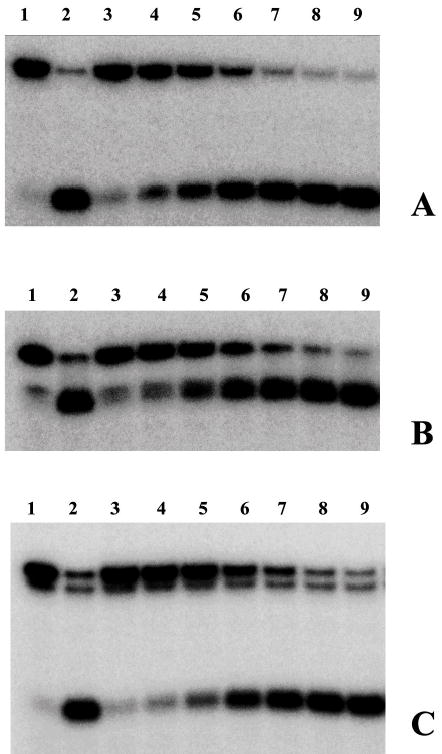
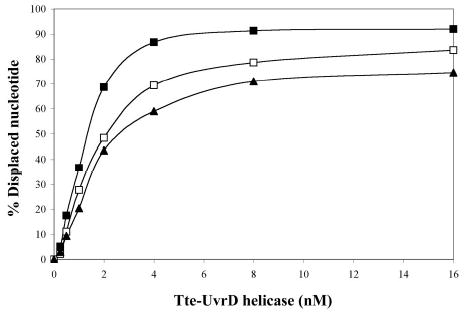
Comparison of the ability of Tte-UvrD protein to unwind DNA duplex with 3′-ssDNA tail, blunt-end, and 5′ -ssDNA tails. Helicase activity assays were performed as described in Materials and Methods. Panel A to panel C are autoradiographs showing the displacement of a 24-mer radio labeled fragment (lower band) from its duplex (upper band). The substrates in panel A were 3′-ssDNA tailed duplexes, in panel B were blunt-ended duplexes, in panel C were 5′-ssDNA tailed duplexes. The DNA concentrations were 0.25nM of duplex DNA molecules. In each panel, lane 1 was the negative control without helicase, lane 2 was the positive control (after heating at 95°C for 15 min without helicase), lanes 3–9 contained Tte-UvrD at 0.25, 0.5, 1, 2, 4, 8, and 16 nM, respectively. The oligonucleotide displacement percentage from panels A to C were calculated and shown in Panel D. Filled square, 3′-ssDNA tailed duplex; open square, blunt-ended duplex; filled triangle, 5′-ssDNA tailed duplex. Each data point represents the result of a single experiment.
