Abstract
A selection strategy, the activator trap, was used in order to identify genes of human cytomegalovirus (HCMV) that encode strong transcriptional activation domains in mammalian cells. This approach is based on the isolation of activation domains from a GAL4 fusion library by means of selective plasmid replication, which is mediated in transfected cells by a GAL4-inducible T antigen gene. With this screening strategy, we were able to isolate two types of plasmids encoding transactivating fusion proteins from a library of random HCMV DNA inserts. One plasmid contained the exon 3 of the HCMV IE-1/2 gene region, which has previously been identified as a strong transcriptional activation domain. In the second type of plasmid, the open reading frame (ORF) UL26 of HCMV was fused to the GAL4 DNA-binding domain. By quantitative RNA mapping using S1 nuclease analysis, we were able to classify UL26 as a strong enhancer-type activation domain with no apparent homology to characterized transcriptional activators. Western blot analysis with a specific polyclonal antibody raised against a prokaryotic UL26 fusion protein revealed that two protein isoforms of 21 and 27 kDa are derived from the UL26 ORF in both infected and transfected cells. Both protein isoforms, which arise via alternative usage of two in-frame translational start codons, showed a nuclear localization and could be detected as early as 6 h after infection of primary human fibroblasts. By performing Western blot analysis with purified virions combined with fractionation experiments, we provide evidence that pUL26 is a novel tegument protein of HCMV that is imported during viral infection. Furthermore, we observed transactivation of the HCMV major immediate-early enhancer-promoter by pUL26, whereas several early and late promoters were not affected. Our data suggest that pUL26 is a novel tegument protein of HCMV with a strong transcriptional activation domain that could play an important role during initiation of the viral replicative cycle.
Human cytomegalovirus (HCMV) is an extensively studied human herpesvirus that can cause a broad spectrum of diseases ranging from subclinical infection in normal healthy individuals to congenital malformation in newborns and life-threatening pneumonitis in immunocompromised adults (6). During lytic infection, more than 200 HCMV genes are expressed in a temporally regulated cascade consisting of three major waves of viral gene expression (8). A few of the encoded proteins have been identified as regulatory molecules that are involved in the coordinate expression of genes during the three distinct phases of the viral replicative cycle, termed the immediate-early (IE), early, and late phases (11, 36, 53, 54). The best-characterized regulatory proteins are the 72-kDa IE1 (UL123) and the 86-kDa IE2 (UL122) polypeptides that originate from the major IE gene region of HCMV during initial stages of the replicative cycle (23, 38, 49). Both IE1-p72 and IE2-p86 are able to positively regulate gene expression independently (35, 50). This has been demonstrated for several heterologous promoters such as the human immunodeficiency virus long terminal repeat (LTR) or the hsp70 promoter (5, 17, 21). In particular, however, a strong cooperative transactivation of viral early promoters has been demonstrated for these two polypeptides (35, 50). A similar cooperative transactivation in combination with major IE proteins was reported for polypeptides derived from three other HCMV gene regions, such as the IE regions UL36-38 and IRS1/TRS1 and the early gene locus UL112/113 (10, 27, 48). In addition, the tegument proteins encoded by the UL69 and UL82 loci are able to exert an independent regulatory function on the major IE enhancer-promoter of HCMV (32, 58).
One characteristic of cellular transcription factors is that their modular structure typically consists of a DNA-binding domain tethering the protein to a particular promoter via sequence-specific protein-DNA interactions and an activation domain for interaction with other factors of the transcriptional apparatus (for a review, see reference 51). Up to now, only for the IE2-p86 protein of HCMV has such a bipartite domain structure been demonstrated: a DNA-binding domain that contacts specific DNA sequences primarily via the minor groove of the DNA helix is located within the C terminus of the protein (9, 30); a transcriptional activation domain with acidic characteristics could be identified within the N-terminal 85 amino acids of the protein (38). An additional activation domain was found within the C-terminal 33 amino acids of IE2-p86; however, the function of this domain appears to be cryptic within the context of the entire protein, since activation could only be observed when this particular domain was fused directly to the GAL4 DNA-binding domain (38).
In an attempt to identify additional HCMV proteins with a function in transcriptional regulation, we used a screening strategy that is able to select strong transcriptional activation domains from a library of random DNA fragments in mammalian cells, the activator trap (13, 19). By using this approach, we were able to select a novel activation domain contained within the open reading frame (ORF) UL26 of HCMV. A further characterization of the encoded protein revealed that pUL26 is a component of the viral tegument that is imported during viral infection and shows a predominantly nuclear localization within infected cells. Furthermore, pUL26 was able to transactivate the major IE enhancer-promoter of HCMV. These results suggest that the UL26 ORF encodes a protein exerting an important function for the initiation of the viral replicative cycle.
MATERIALS AND METHODS
Library construction and activator trap assay.
A total of 50 μg of a mixture of cosmid DNAs (pCM1052, pCM1050, pCM1058, pCM1007, pCM1029, pCM1049, pCM1015, and pCM1035) (14) covering the entire HCMV strain AD169 genome was sonicated by using the Branson Sonifier System. Sonicated fragments of ca. 500 bp were selected by electrophoresis in a 1% agarose gel and repaired by treatment with T4 polymerase, Klenow polymerase, and T4 kinase. The plasmid library was constructed by ligating the sonicated DNA fragments into the EcoRV site of the GAL4 expression plasmid pSCTEV-GAL4(1-93)RV and by transformation into Escherichia coli DH1 by electroporation (19). The following steps of the activator trap assay (isolation of library DNA, transfection of CV-1-5GT cells, extraction of low-molecular-weight DNA from transfected cells, digestion with DpnI, and electroporation of competent DH1 E. coli cells) were performed exactly as described previously (19). In order to confirm the transactivating function of the selected GAL4 fusions, plasmid DNA of individual ampicillin resistant clones was isolated and used for cotransfection of HeLa cells, together with a luciferase reporter plasmid bearing five GAL4 sites upstream of the adenovirus E1b promoter. GAL4 fusion clones containing transcriptional activation domains were sequenced by the Sanger dideoxynucleotide technique by using a commercially available T7 sequencing kit (43).
Construction of plasmids.
Construction of the GAL4 expression vector pSCTEV-GAL4(1-93)RV and the β-globin reporter genes 5GOVEC, p2Sp1/β5G-OVEC, and OVEC-1 have been described previously (19, 44, 56). Plasmid pHM246, containing the UL26 gene region, was constructed by cleavage of cosmid pCM1017 (14) with SmaI and SalI and insertion of the respective 1-kb fragments into the pBluescript KSII vector (Stratagene, Amsterdam, The Netherlands). To obtain a probe for Northern blot analysis, the UL26 ORF was amplified by PCR with the primer pair UL26up (5′-CAGAGGATCCCGATGACGAGTAGGCGCG-3′) and UL26ds (5′-ATCTGGATCCTTACGGCAACAGCGCTGA-3′) and plasmid pHM246 as the template. The respective fragment was cleaved with BamHI and inserted into the pBluescript KSII vector (Stratagene), resulting in plasmid pHM281. Prokaryotic expression vector pHM312 expressing UL26 as a His-tagged protein was created via PCR amplification of the UL26 coding sequence with the primer pair UL26up and UL26ds-HindIII (5′-ATCTAAGCTTTTACGGCAACAGCGCTGA-3′), followed by cleavage with BamHI and HindIII and ligation into the pQE32 vector (Qiagen). For construction of a eukaryotic expression vector for UL26, plasmid pHM246 was cleaved with XbaI and SalI, followed by a filling-in reaction with Klenow enzyme and isolation of the genomic UL26 fragment via gel purification. The respective fragments were then inserted into the eukaryotic expression vector pCB6 which had been cleaved with KpnI and treated with Klenow enzyme. A second series of UL26 expression vectors (pHM619 and pHM620) was created by inserting the PCR fragment that had been used for construction of plasmid pHM312 in either a sense or an antisense orientation into the eukaryotic expression vector pCB6 that was cleaved either with BglII and HindIII or with BamHI and HindIII, respectively. Plasmids pHM1773 and pHM1774, which were used as templates in in vitro transcription-translation reactions, were constructed as follows. UL26 was amplified via PCR with either the primer pair UL26ATG1 (5′-ATGCGGATCCATGTACGCCGTTTTCGGCC-3′)-UL26pcDNA3 (5′-ATGCGATATCTTACGGCAACAGCGC-3′) or the primer pair UL26ATG2 (5′-ATGCGGATCCATGACGAGTAGGCGCGC-3′)-UL26pcDNA3. The resulting fragments were cleaved with BamHI and EcoRV, followed by insertion into the pcDNA3 vector (Invitrogen, Karlsruhe, Germany). Luciferase reporter plasmids pHM287 (major IE enhancer promoter), pHM142 (UL112/113 promoter), pHM571 (UL84 promoter), and pHM145 (UL86 promoter) were described previously (24, 57, 58).
Cell culture, virus infections, transfections, and luciferase assays.
CV-1-5GT cells were maintained in minimal essential medium (Gibco-BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum. For the activator trap assay, this cell line was transfected with 100 ng of library DNA per 100-mm dish by using the DEAE-dextran method (16). HeLa, U373, and COS-7 cells and primary human foreskin fibroblasts (HFFs) were cultured and transfected by calcium phosphate coprecipitation with BES and by using the Fugene transfection reagent as described previously (2, 24, 58). For the establishment of stably transfected U373 cell lines, G418 (Gibco-BRL) was added 48 h after transfection at a concentration of 500 μg/ml. Four weeks later, individual colonies were isolated and analyzed for UL26 protein expression by indirect immunofluorescence. Infection of HFF cells with HCMV (AD169) and treatment of cells with cycloheximide (CHX) or phosphonoformic acid were performed exactly as described previously (40, 47). Luciferase assays were also performed as described previously (24).
RNA extraction and S1 analysis.
About 48 h after transfection, the total cellular RNA was harvested as described previously (47). For S1 nuclease analysis a single-stranded oligonucleotide probe extending between positions −18 and +75 of the rabbit β-globin gene was labeled by using [γ-32P]ATP and polynucleotide kinase (28). RNA (20 μg) was hybridized with the probe (50,000 cpm) overnight at 30°C in 10 μl of hybridization buffer containing 80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.4], 400 mM NaCl, and 1 mM EDTA. Digestion of single-stranded RNA and single-stranded DNA was performed for 1 h at room temperature with 150 U of S1 nuclease in S1 buffer containing 0.25 M NaCl, 30 mM sodium acetate, 1 mM ZnSO4, 13 μg of herring sperm DNA/ml, and 10 μg of single-stranded calf thymus DNA/ml. After phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation, the samples were analyzed in a 10% polyacrylamide-7.5 M urea gel, followed by autoradiography of the dried gels.
Northern (RNA) blot analysis.
For Northern blot analysis, total cellular RNA harvested at the indicated time points was separated by using 1.2% formaldehyde agarose gels as described previously (47). The RNA was transferred to Hybond-N membranes (Amersham, Braunschweig, Germany), fixed to the filters by heat (2 h, 80°C), and prehybridized for at least 4 h at 43°C in prehybridization solution containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50 mM sodium phosphate (pH 6.5), 5× Denhardt solution, and 1 mg of yeast RNA/ml. As radioactive probes, double-stranded DNA fragments corresponding either to the entire UL26 ORF or to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were labeled by nick translation as described previously (47). Hybridizations were performed for 24 h at 43°C in hybridization solution containing 50% formamide, 5× SSC, 20 mM sodium phosphate (pH 6.5), 1× Denhardt solution, 500 μg of yeast RNA/ml, and the radioactive probe at 500,000 cpm/ml. The filters were washed several times after hybridization with buffer containing 20 mM sodium phosphate, 0.1% sodium dodecyl sulfate (SDS), and decreasing concentrations of SSC. For reprobing of the filters the radioactive probe was removed by incubation for 3 h at 65°C in a buffer containing 5 mM Tris-HCl (pH 8.0), 2 mM EDTA (pH 8.0), and 0.1× Denhardt solution with several changes of the buffer.
Protein expression and purification, immunization, immunoblotting, and indirect immunofluorescence analysis.
Prokaryotic expression and purification of UL26 as a histidine-tagged protein were performed as described previously (29). The isolated protein was dialyzed against 1% SDS and stored in aliquots at −80°C. In order to generate specific antisera, rabbits were injected intramuscularly with 200 μg of protein five times and bled after ca. 4 months (performed by Eurogentec, Seraing, Belgium). In vitro-translated proteins were generated by using the TNT system (Promega, Heidelberg, Germany) as described previously (58). Purification of HCMV virions and dense bodies and detergent treatment of infectious virions were performed as described previously (60). Immunoblotting was performed essentially as described previously (58). Briefly, proteins were separated by SDS-15% polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred from acrylamide gels to nitrocellulose filters. In order to diminish nonspecific reactions, rabbit antisera were preincubated with total E. coli lysate that had been transferred to nitrocellulose filters. Blocking of filters and immunostaining were done either as described or by using ECL reagents (Amersham) (58). For indirect immunofluorescence, cells were fixed with methanol, and a 1:200 dilution of the rabbit antisera was layered over the cells for 30 min at 37°C. Nonspecific binding as observed with infected HFFs was blocked by preincubation with 12.5% horse serum for 1 h at 37°C. Fluorescein-conjugated anti-rabbit immunoglobulin (Dako GmbH, Hamburg, Germany) was added for 30 min (1:40 dilution). After each incubation step, cells were washed extensively with phosphate-buffered saline.
RESULTS
Selection of strong transcriptional activation domains from the HCMV genome by the activator trap assay.
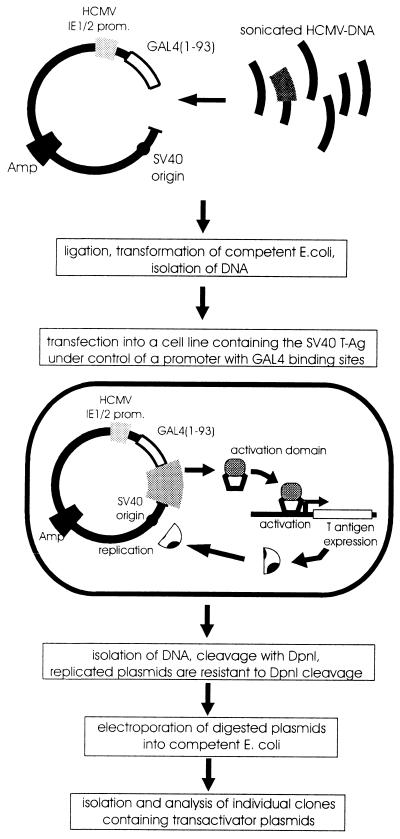
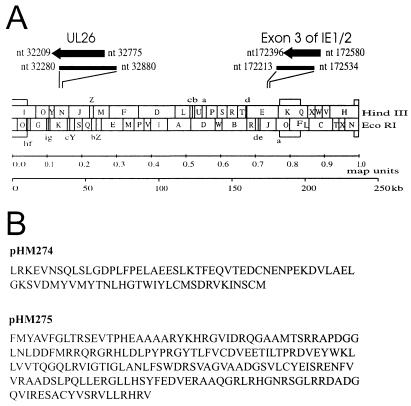
In order to identify novel proteins of HCMV with a potential role in transcriptional regulation, we used a screening strategy that is able to select strong transcriptional activation domains from a library of random DNA inserts in mammalian cells (19). This approach is based on a cell line containing the large T-antigen of simian virus 40 (SV40) under control of a promoter with binding sites for the yeast factor GAL4. Transfected plasmids that express functional transactivating fusion proteins with the GAL4 DNA-binding domain are able to activate T-antigen expression, thus promoting selective amplification of the introduced plasmids via the SV40 origin of replication that is contained within each plasmid (see Fig. 1). In order to utilize this procedure for HCMV, DNA comprising the entire genome of HCMV, strain AD169, was sonicated to fragments of ca. 500 nucleotides (nt) and then cloned into the eukaryotic expression vector pSCTEV-Gal4(1-93)RV, downstream of the DNA-binding domain of the yeast factor GAL4. DNA from this expression library, consisting of randomly generated plasmids of HCMV fragments fused to the GAL4 DNA-binding domain, was transfected in pools of ca. 500 different clones per 100-mm plate into the monkey kidney cell line CV-1-5GT, containing the large T antigen of SV40 under control of a promoter with five GAL4-binding sites. The introduction of a plasmid encoding a functional transactivator protein was then able to stimulate T-antigen expression, thus leading to a selective replication of the respective construct. After extraction of DNA from transfected cells, replicated plasmids were isolated via digestion with DpnI that selectively degrades nonreplicated plasmids and the residual DNA was used for transformation of E. coli cells. The transactivating function of individual GAL4 fusion clones isolated by this screening assay was confirmed by cotransfection with a GAL4-dependent luciferase reporter plasmid and subsequent quantification of luciferase activity (data not shown). By using this procedure we were able to select two types of transactivating GAL4 fusion clones in multiple copies as the only clones with strong transactivation capacity. Nucleotide sequence analysis of the respective inserts revealed that one type of clones (termed pHM274) contained 139 bp of the exon 3 of the major IE gene region of HCMV (see Fig. 2); this domain has previously been described as a strong transcriptional activation domain of the IE2 transactivator protein, thus confirming the functionality of our screening strategy (38). The 601-bp insert of the second type of clones (termed pHM275) corresponded in sequence to nearly the entire ORF UL26 of HCMV (see Fig. 2). This reading frame of 567 nt, which is located in the left part of the UL gene region, can code for a protein of 188 amino acids (predicted molecular mass of 21 kDa) of unknown function.
FIG. 1.
Selection of activation domains from the HCMV genome by the activator trap assay. DNA from cosmid clones representing the entire HCMV strain AD169 genome was fragmented by sonication and fragments of ca. 500 bp were cloned into the expression vector pSCTEV-GAL4(1-93)RV downstream of the DNA-binding domain of the yeast factor GAL4. DNA isolated from the GAL4 fusion library was transfected into CV-1-5GT cells with the SV40 T-antigen under the control of five GAL4-binding sites. Expression of the transactivating GAL4 fusion protein stimulated the production of SV40 T antigen, thereby leading to replication of the SV40 origin-containing library plasmids. After extraction of DNA, replicated plasmids were isolated by cleavage with DpnI. E. coli cells were transformed with nondegraded plasmids, which allowed subsequent isolation of the individual clones encoding activation domains.
FIG. 2.
Prototype arrangement of the HCMV AD169 genome and positions of DNA fragments (A) and amino acid sequence of activation domains (B) isolated by the activator trap system. (A) The prototype arrangement of the HCMV AD169 genome is shown in the lower half. In the upper half the position of the HCMV DNA fragments isolated by the activator trap assay and the positions of exon 3 of the IE1/2 gene region and the ORF UL26 are indicated (the numbers refer to the prototype sequence of the HCMV AD169 genome). (B) Amino acid sequence of activation domains as contained within plasmid pHM274 (exon 3 of IE1/2) or pHM275 (UL26).
The ORF UL26 contains a transcriptional activation domain that is able to stimulate transcription from both promoter-proximal and promoter-remote positions.
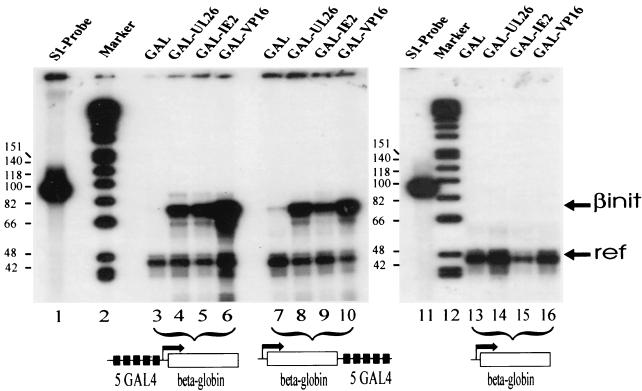
In order to confirm the transactivation capacity of the selected domains, we decided to perform a quantitative RNA mapping by using an S1 nuclease analysis. Transcriptional activation domains can be functionally classified into at least two types (44): glutamine-rich domains only stimulate transcription when positioned in a promoter position near the initiation site, whereas enhancer-type activation domains can activate transcription from remote and proximal positions. The latter type is exemplified by the acidic domains contained within the herpes simplex virus VP16 protein. Since examination of the selected UL26 sequence did not reveal known features of proline-, glutamine-, and serine/threonine-rich or acidic activation domains (see Fig. 2B), we wanted to classify whether it was promoter-like or enhancer-like. Therefore, we performed cotransfections of HeLa cells by using two different β-globin reporter constructs with GAL4-binding sites either immediately upstream of the β-globin promoter (plasmid p5G-OVEC) or in a remote position downstream of the reporter gene (plasmid p2Sp1/β5G-OVEC). As a control for nonspecific activation, a β-globin reporter plasmid without GAL4-binding sites was used (plasmid OVEC-1). In addition, a reference plasmid was transfected, which gives rise to a truncated β-globin transcript and allows for normalization of transfection variations (56). After isolation of RNA and quantitative S1 nuclease analysis, the result shown in Fig. 3 could be observed. Cotransfection of a promoter-proximal reporter plasmid with the GAL4-UL26 expression construct resulted in a strong signal that was weaker than the signal observed in the presence of a GAL4-VP16 fusion protein but comparable to RNA levels obtained after cotransfection of the GAL4-IE2 fusion that was also isolated in the activator trap assay (see Fig. 3, lanes 3 to 6). No stimulation of β-globin transcription was seen with a reporter construct without GAL4-binding sites, confirming the specificity of the observed effects (Fig. 3, lanes 13 to 16). When the reporter plasmid with GAL4-binding sites in a remote position was used, transactivation by GAL4-UL26 was not diminished, demonstrating a propensity of UL26 to activate from a remote position (Fig. 3, lane 8). Thus, the UL26 ORF encodes a strong transcriptional activation domain that can stimulate transcription from both promoter-proximal and promoter-remote positions.
FIG. 3.
The selected UL26 activation domain is active both from a promoter-proximal position and from a remote enhancer position. HeLa cells were transfected with 10 μg of β-globin reporter plasmids, 1 μg of expression vectors encoding the GAL4 fusion proteins, and 1 μg of the reference plasmid OVEC-Ref. β-Globin RNA transcribed from the reporter plasmids was isolated from HeLa cells, hybridized to a 93-nt S1 probe covering position −18 to position +75 of a noncoding region of the β-globin reporter gene and mapped by using S1 nuclease. “βinit” indicates the correctly initiated transcripts of the reporter gene, and “ref” indicates transcripts of the reference plasmid OVEC-Ref, which was cotransfected as a control for transfection variation. Lanes: 1 and 11, S1 nuclease probe; 2 and 12, molecular mass markers (sizes [in nucleotides] are indicated on the left); 3 to 6, transfection with a β-globin reporter plasmid containing five GAL4-binding sites upstream of a minimal β-globin promoter (promoter proximal position); 7 to 10, transfection with a β-globin reporter plasmid containing five GAL4-binding sites downstream of the β-globin gene (remote position); 13 to 15, transfection with a β-globin reporter plasmid without GAL4-binding sites. In lanes 3, 7, and 13, cotransfection was performed with a vector expressing the GAL4 DNA-binding domain. In lanes 4, 8, and 14, cotransfection was performed with plasmid pHM275, encoding a GAL4-UL26 fusion protein. In lanes 5, 9, and 15, cotransfection was performed with plasmid pHM274, encoding a fusion of exon 3 sequences of IE2 to GAL4. In lanes 6, 10, and 16, cotransfection was performed with a plasmid encoding a fusion of GAL4 to the herpes simplex virus type 1 VP16 activation domain, which served as a positive control.
Analysis of transcripts from the UL26 gene region reveals an early-late kinetics of expression.
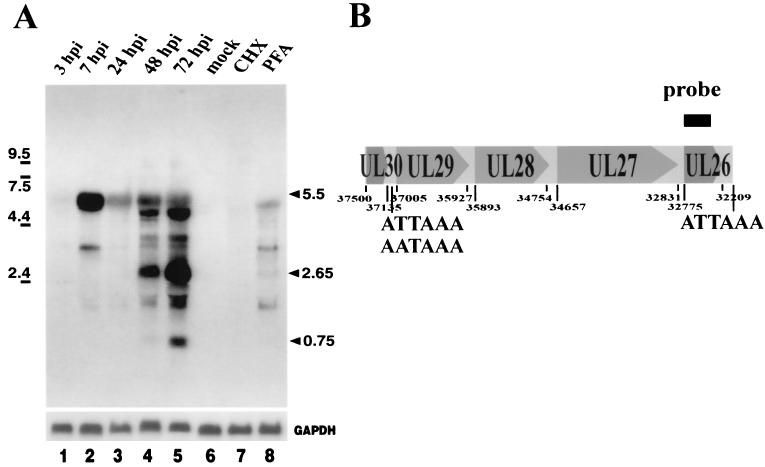
Since no data on expression from the UL26 gene region were available, we decided to analyze the pattern of transcripts first. In order to investigate the time course of RNA expression from the UL26 gene locus, RNA from HCMV-infected fibroblast cells was harvested. To obtain a probe for Northern blot experiments, the UL26 gene region was amplified by PCR and cloned into the pBluescript KSII vector. Northern blot experiments were then performed to determine the size(s) of transcripts from UL26. Total cellular RNA, harvested at the indicated time points, was separated on 1.2% formaldehyde agarose gels, transferred to nylon filters, and hybridized with a probe covering the entire UL26 ORF. As shown in Fig. 4, we observed a complex pattern of transcripts that changed during the time course of the HCMV replicative cycle. Weak transcription was detected as early as 3 h after infection and increased dramatically in abundance at 7 h postinfection (hpi). However, no significant signal was present in RNA harvested in the presence of CHX, arguing against an IE gene. Whereas during early times a transcript of 5.5 kb dominated, we could observe an abundant transcript of 2.65 kb and a smaller transcript of 0.75 kb that were first detectable at 48 h after infection. In summary, these results demonstrate a complex expression of transcripts from UL26 that appear with early-late kinetics during the replicative cycle of HCMV.
FIG. 4.
Northern blot analysis with RNA isolated during the HCMV replicative cycle after infection of primary HFFs. (A) A probe specific for the UL26 ORF was used for hybridization (see panel B). The lower part of panel A shows a control hybridization with a probe specific for GAPDH mRNA. Lanes: 1 to 5, 10 μg of RNA harvested at 3, 7, 24, 48, and 72 hpi, respectively; 6, 10 μg of RNA harvested from uninfected cells; 7, 10 μg of RNA harvested at 6 hpi in the presence of CHX (150 μg/ml); 8, 10 μg of RNA harvested at 72 hpi in the presence of phosphonoformic acid (200 μg/ml). The sizes of molecular weight markers are indicated on the left; the arrows on the right show dominant transcripts. (B) Schematic diagram of the UL30-UL26 genomic region. The location of ORFs with nucleotide positions of start and stop codons and the relative positions of potential polyadenylation signals are indicated.
Prokaryotic expression of UL26 and generation of a specific antiserum.
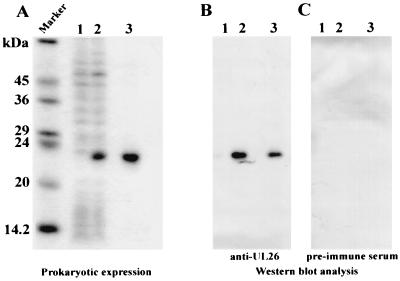
In order to obtain protein for the generation of a specific antiserum, the UL26 ORF was expressed in E. coli. We decided to express UL26 as a histidine-tagged protein, since this system allows for very efficient purification under denaturing conditions of even small amounts of expressed protein via metal chelate affinity chromatography. For this purpose, the UL26 ORF was amplified by PCR and inserted into the prokaryotic expression vector pQE32, which resulted in an in-frame fusion to 13 vector-encoded amino acids at the N terminus, including the histidine tag. Expression from this construct was analyzed by evaluating the induction of protein synthesis in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside), followed by SDS-PAGE and Coomassie blue staining of the gel. As can be seen in Fig. 5A, lanes 1 and 2, there was a strong induction of a protein of 23 kDa in the presence of IPTG. In parallel, the protein was purified via metal chelate affinity chromatography as described in Materials and Methods (Fig. 5A, lane 3). The purified protein was then used to immunize rabbits. The resulting rabbit serum against the UL26 fusion protein was tested for its specificity by Western blot analysis. As shown in Fig. 5B, the specific antiserum exclusively detected the UL26 fusion protein within both total cellular lysates and purified fractions of induced E. coli cells, whereas no specific reaction could be observed with proteins from uninduced cells or with the use of preimmune serum. Thus, a specific antiserum could be generated for a further analysis of protein expression from UL26.
FIG. 5.
Affinity purification of prokaryotically expressed, His-tagged UL26 protein. Results obtained by 15% PAGE with Coomassie blue stain (A) and Western blot analysis with specific anti-UL26 antiserum (B) and preimmune serum (C) are shown. Lanes: 1, cell lysate from uninduced cells; 2, cell lysate from IPTG-induced cells; 3, affinity-purified protein from IPTG-induced cells. Lane M shows the molecular mass marker, with the sizes indicated on the left.
Eukaryotic expression analysis of UL26 protein after transfection and infection of cells.
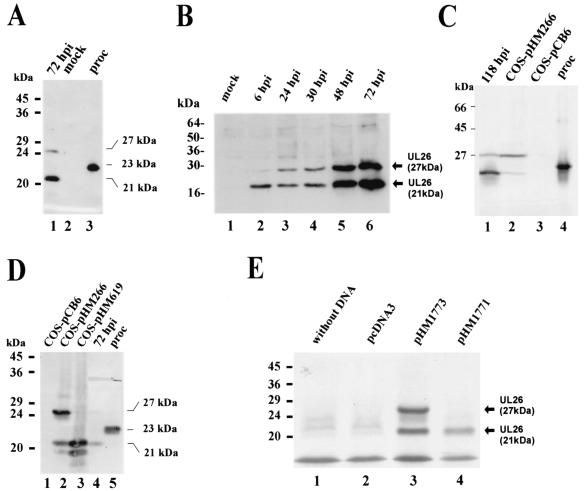
Western blot analyses were performed in order to detect pUL26 in HCMV-infected cells. In a first series of experiments, lysates from HCMV-infected cells at 72 hpi were separated, together with lysates from mock-infected cells, and prokaryotically expressed pUL26 by SDS-PAGE. After transfer to nitrocellulose filters and reaction with the specific UL26-antiserum, we could observe a strong signal corresponding to a protein of 21 kDa and a second weaker band at 27 kDa with infected cell extracts (Fig. 6A, lane 1). No signal was obtained with lysates from mock-infected cells (Fig. 6A, lane 2). As observed in previous experiments, the prokaryotically expressed pUL26 that was modified by the addition of an N-terminal His tag migrated at 23 kDa (Fig. 6A, lane 3). To further analyze the kinetics of pUL26 expression during the viral replicative cycle, cell lysates were harvested at various time points after infection with HCMV and examined by immunoblotting with the UL26 antiserum. Figure 6B shows the results that we obtained. A specific band at 21 kDa was first detected at 6 hpi. The expression increased at later times, when the additional 27-kDa protein could also be observed. Interestingly, when lysates from COS-7 cells that had been transfected with a eukaryotic expression vector for UL26 (termed pHM266) were investigated in parallel, both the 21-kDa and the 27-kDa forms of pUL26 were present; however, in contrast to infected cells, the upper protein was more abundant (Fig. 6C, lanes 1 and 2). These results suggested that pUL26 might undergo posttranslational modification in eukaryotic cells. In order to investigate this, we performed immunoprecipitation analysis of 32P- and 35S-labeled infected cell extracts with the UL26 antiserum. This resulted in the detection of both the 21- and the 27-kDa proteins after 35S labeling; however, no signal was obtained with 32P-labeled cell extracts, suggesting that pUL26 is not modified by phosphorylation (data not shown). On a closer inspection of the sequence located upstream of the UL26 ORF, as predicted by Chee et al. (8), we could detect an in-frame ATG located 34 amino acids 5′ of the predicted ATG (8). This second ATG is also included in the coding sequence contained within the eukaryotic UL26 expression vector pHM266. In order to test the hypothesis that usage of an upstream in-frame ATG could give rise to the 27-kDa UL26 protein, we constructed an additional eukaryotic expression vector without the upstream ATG, termed pHM619. After transfection of this vector into COS-7 cells and analysis of the expressed proteins by immunoblotting, we were no longer able to detect the 27-kDa UL26 protein, suggesting that the two isoforms of pUL26 can be explained by usage of two in-frame start codons (see Fig. 6D). To further confirm this hypothesis, in vitro transcription-translation reactions were performed with plasmids pHM1773 and pHM1774 as templates. Plasmid pHM1773 contained both ATGs that were detected within UL26, whereas the coding sequence of pHM1774 started at the predicted ATG. As can be seen in Fig. 6E, in vitro transcription-translation reactions with pHM1773 resulted in two proteins of 21 and 27 kDa, whereas only one protein (21 kDa) could be detected when pHM1774 was used as a template (Fig. 6E, lanes 3 and 4).
FIG. 6.
Eukaryotic expression analysis of the UL26 protein after infection of HFFs with HCMV strain AD169 or after transfection of COS-7 cells. (A) Western blot analysis with specific anti-UL26 antiserum and cell extracts from HFFs harvested at 72 h after infection with HCMV. Lanes: 1, extracts from HFFs 72 hpi with HCMV; 2, extracts from mock-infected HFFs; 3, prokaryotically expressed UL26 protein. (B) Western blot analysis with anti-UL26 antiserum and protein extracts harvested at various times after infection of human fibroblast cells with HCMV. Lanes: 1, mock-infected cells; 2, 6 hpi; 3, 24 hpi; 4, 30 hpi; 5, 48 hpi; 6, 72 hpi. (C) Western blot analysis with anti-UL26 antiserum and protein extracts from infected HFFs and transfected COS-7 cells. Lanes: 1, extracts from HFFs, 118 hpi; 2, extracts from COS-7 cells transfected with UL26 expression vector pHM266; 3, extracts from COS-7 cells transfected with expression vector pCB6; 4, prokaryotically expressed UL26. (D) Western blot analysis with anti-UL26 antiserum and extracts from transfected COS-7 cells. Lanes: 1, extracts from COS-7 cells that were transfected with the vector pCB6; 2, extracts from COS-7 cells that were transfected with UL26 expression plasmid pHM266; 3, extracts from COS-7 cells that were transfected with UL26 expression plasmid pHM619; 4, extracts from HFFs at 72 hpi with HCMV; 5, prokaryotically expressed pUL26. (E) Analysis of in vitro-translated proteins by SDS-PAGE and autoradiography. Lanes: 1, no template was used in the in vitro transcription-translation reaction; 2, the vector pcDNA3 was used in the in vitro transcription-translation reaction; 3, plasmid pHM1773 (containing both ATGs of UL26) was used in the in vitro transcription-translation reaction; 4, plasmid pHM1774 (containing only the predicted ATG of UL26) was used in the in vitro transcription-translation reaction. Sizes of the molecular mass markers are shown on the left of each panel; the sizes of pUL26 proteins are indicated on the right of panels A, B, D, and E.
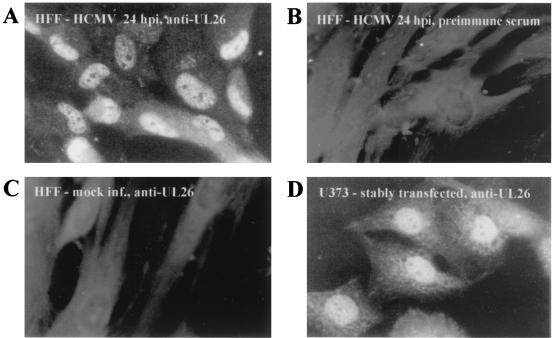
To determine the subcellular localization of pUL26, indirect immunofluorescence analysis of both infected and stably transfected cells was performed. HFFs that were infected for 24 h with HCMV AD169 were incubated with the specific UL26 antiserum and then stained with fluorescein-conjugated anti-rabbit antibodies. This revealed a predominantly nuclear staining of infected HFFs with exclusion of the nucleoli (Fig. 7A). The same pattern of strong nuclear and weak cytoplasmic staining was also observed with U373 cells that stably expressed the UL26 protein, showing that no other viral protein is required for nuclear localization of UL26 (Fig. 7D). No signals could be detected with mock-infected cells or when preimmune serum was used for staining (Fig. 7B and C). In summary, these results show that two proteins of 21 and 27 kDa with a predominantly nuclear localization originate from the UL26 ORF with an early-late kinetics of expression.
FIG. 7.
Immunofluorescence analysis of HCMV-infected HFFs and stably transfected U373 cells. Specific antiserum was used for the staining of HCMV-infected (24 hpi) or mock-infected HFFs (A and C, respectively) or stably transfected U373 cells (D). No specific signal could be observed when HCMV-infected HFFs (24 hpi) were probed with the preimmune serum (B).
Identification of pUL26 as a novel tegument protein of HCMV.
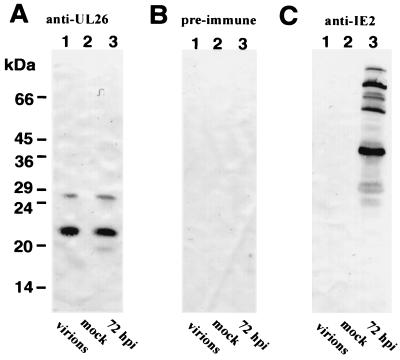
In a previous publication by Baldick and Shenk (4), the UL26 protein has been reported to be associated with purified HCMV particles (4). In order to confirm and extend these findings, we sought to determine whether we could detect pUL26 within infectious virions with our specific anti-UL26 antiserum. To investigate this, virions were isolated via density gradient centrifugation, and Western blot analyses were performed in order to detect pUL26. Cell extracts from infected (at 72 hpi) and uninfected fibroblasts were used in parallel as positive and negative controls, respectively. As shown in Fig. 8A, lanes 1 to 3, the rabbit serum against pUL26 could detect UL26 proteins in virions and HCMV-infected cells but not in uninfected cells. No specific reactivity was seen with a preimmune serum (Fig. 8B). To assess the purity of the virion preparation, we did immunoblot analysis with a rabbit serum directed against IE2 proteins of HCMV (15). Several IE2 polypeptides are expressed during the late phase of the HCMV replicative cycle but are not contained within virions (39). As can be seen in Fig. 8C, several IE2 proteins could be detected in infected fibroblasts but not in virions or in uninfected cells. This excludes the possibility that the pUL26 signal in virions is due to a contamination of the virion preparation with cellular material.
FIG. 8.
Detection of UL26 protein of HCMV in purified virus particles by Western blot analysis. Western blot analysis with specific anti-UL26 antiserum (A), preimmune serum (B), or IE2-specific rabbit antiserum (C). Lanes: 1, purified HCMV particles; 2, extracts from mock-infected fibroblast cells; 3, extracts from HCMV-infected fibroblast cells (at 72 hpi). Proteins were separated by SDS-15% PAGE. The sizes of molecular mass markers are shown on the left.
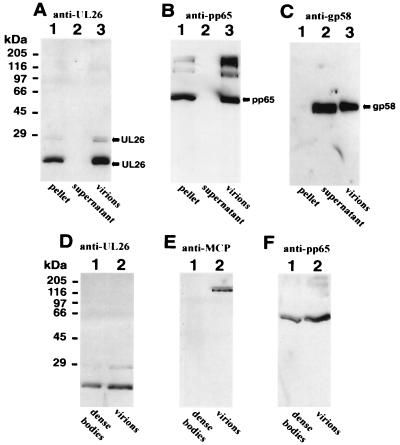
We next wanted to determine the localization of pUL26 within the virus particle. First, a detergent treatment of purified virions was performed; this should result in a separation of capsid and tegument proteins from polypeptides contained within the envelope. After incubation of virions in the presence of the detergents NP-40 and deoxycholate, the viral material was centrifuged in order to pellet the insoluble capsid and/or tegument proteins. Afterwards, polypeptides contained within the supernatant and pellet were separated via SDS-PAGE. In order to assess the purity of the obtained fractionation, we performed Western blot experiments with monoclonal antibodies directed against typical envelope or tegument proteins. As shown in Fig. 9, the tegument protein pp65 could exclusively be detected in the pellet fraction (Fig. 9B, lane 1), whereas the glycoprotein gB was only contained within the supernatant fraction (Fig. 9C, lane 2). Using the specific rabbit antiserum directed against pUL26, we found that both forms of the UL26 protein cofractionated with the capsid or tegument proteins in the pellet (Fig. 9A, lane 1). Therefore, pUL26 is not associated with the virion envelope but, instead, is located within the rather tightly associated capsid or tegument structure. To resolve the question as to whether UL26 proteins are located within the tegument or the capsid, we made use of the observation that dense bodies, which are defective viral particles produced during high-multiplicity infection of fibroblasts, are devoid of capsid proteins (25, 26, 31). HCMV virions and dense bodies were purified via two consecutive gradient centrifugations. After separation of the proteins contained within virions and dense bodies via SDS-PAGE, the protein composition was investigated by immunoblot analysis. Using a monoclonal antibody directed against the major capsid protein of HCMV, we could only detect a signal within the virion fraction (Fig. 9E, lane 2). This demonstrated that the dense body fraction was almost free of virions (Fig. 9E, lane 1). After removal of the monoclonal antibody against the major capsid protein by incubation in a buffer containing 1% SDS, the same filter was reprobed with the rabbit serum directed against pUL26. As shown in Fig. 9D, pUL26 could be detected both in virions and in dense bodies. Therefore, the UL26 protein is not located in the HCMV capsid structure. On the basis of these results, we conclude that pUL26 is a novel tegument protein of HCMV.
FIG. 9.
Identification of pUL26 as a constituent of the tegument. (A to C) Detection of various HCMV polypeptides by Western blot analysis after detergent treatment of purified virus particles and separation of solubilized and nonsolubilized proteins. (A) Western blot analysis with specific anti-UL26 antiserum; (B) monoclonal antibody 28-77 (7) against the tegument protein pp65; (C) monoclonal antibody 27-156 (46) against the glycoprotein gB. Lanes: 1, insoluble proteins after detergent treatment of virions; 2, solubilized proteins after detergent treatment of virions; 3, complete virions that were used for detergent treatment. (D to F) Western blot analysis of dense bodies and virions. (D) Western blot analysis with specific anti-UL26 antiserum; (E) monoclonal antibody 28-4 (42) against the major capsid protein of HCMV; (F) monoclonal antibody 28-77 against the tegument protein pp65. The sizes of molecular mass markers are shown on the left.
UL26 protein import by viral infection.
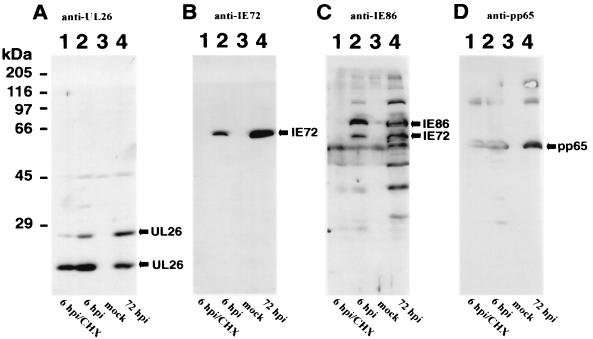
In order to investigate whether pUL26 is present in the IE phase due to uptake from virus inoculum, HFF cells were cultured in the presence of CHX (150 μg/ml) starting at 30 min before and extending treatment for 6 h after infection with HCMV. Cell extracts harvested at 6 hpi in the presence or absence of CHX treatment were separated by SDS-PAGE and analyzed by immunoblotting with polyclonal sera against UL26, IE2-p86, or monoclonal antibodies for the detection of the IE1-p72 and pp65 proteins. As shown in Fig. 10B and C, the IE proteins IE1-p72 and IE2-p86 could only be detected in the absence of CHX, thus demonstrating the efficacy of inhibition of de novo protein synthesis. In contrast, strong signals could be observed for pUL26 and for the known tegument protein pp65 in lysates harvested in the presence of CHX (Fig. 10A and D, lanes 1). Thus, pUL26 is present during the IE phase of the viral replication cycle due to protein import by the viral inoculum.
FIG. 10.
Protein import of pUL26 by viral infection. Western blot analysis of extracts from HFFs that were mock infected or infected with HCMV in the absence or presence of CHX. (A) Anti-UL26 antiserum was used for the detection of UL26 proteins. (B) Monoclonal antibody p63-27 (1) was used for detection of the IE protein IE1-p72. (C) Anti-IE86 antiserum (15) was used for the detection of IE1-p72 and IE2-p86, as well as additional IE2 isoforms. (D) Monoclonal antibody 28-77 was used for the detection of the pp65 tegument protein. Lanes: 1, extracts from HFFs at 6 hpi with HCMV in the presence of CHX; 2, extracts from HFFs at 6 hpi without CHX; 3, extracts from uninfected HFFs; 4, extracts from HFFs at 72 hpi.
Transactivation of the HCMV IE1/2 enhancer-promoter by pUL26.
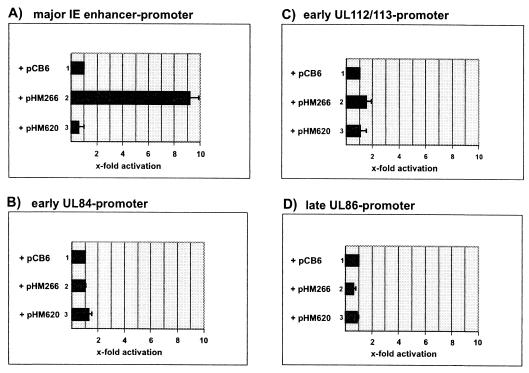
The identification of a strong transcriptional activation domain within the UL26 ORF suggested that pUL26 might be able to transactivate viral promoters. In order to test this hypothesis, transient-expression assays were performed with HFFs. For this, luciferase reporter plasmids containing either the major IE enhancer-promoter of HCMV, the early promoters of the UL112/113 and UL84 gene regions, or the late promoter driving expression of the gene encoding the major capsid protein (UL86) were cotransfected with the UL26 expression vector pHM266. As controls, cotransfections were performed with the empty expression vector pCB6 and plasmid pHM620 which contained the coding sequence of UL26 in antisense orientation. As shown in Fig. 11C, B, and D, no significant effect of pUL26 could be observed on the early or late promoters of the UL112/113, UL84, and UL86 gene regions, respectively. In contrast, however, ∼9-fold activation of the major IE enhancer-promoter could be detected in the presence of pUL26 (Fig. 11A, lane 2). These results suggest that pUL26 is able to act as a specific transactivator of viral IE gene expression.
FIG. 11.
Luciferase analysis after cotransfection of HFFs with various luciferase constructs and UL26 expression plasmids. (A to D) Schematic diagrams of activation values obtained in cotransfection experiments. The relative luciferase activity is expressed as the fold activation relative to the activity of the respective luciferase construct in the absence of an expression vector. The results are from at least three independent experiments; standard deviations are indicated by error bars. Luciferase reporter plasmids containing the HCMV IE1/2 enhancer-promoter (A), the UL112/113 promoter (B), the UL84 promoter (C), or the UL86 promoter (D) were used in cotransfection experiments. Lanes: 1, the empty expression plasmid pCB6 was used for cotransfection; 2, the UL26 expression plasmid pHM266 was used for cotransfection; 3, plasmid pHM620, containing the UL26 gene region in antisense orientation, was used for cotransfection.
DISCUSSION
In this study, we analyzed gene expression from the ORF UL26 of HCMV which was identified in a screen for eukaryotic transactivator proteins. This reading frame has been described to be 567 nt in length and to encode a 188-amino-acid protein with an estimated molecular mass of 21 kDa (8). A search of the Conserved Domain Database at the National Center for Biotechnology Information (Washington, D.C.) revealed a previously unnoted significant similarity of the UL26 ORF to the US22 family of homologous proteins. The US22 genes of CMVs are members of a multigene family unique to the betaherpesviruses that code for proteins involved in the regulation of gene expression (TRS1/IRS1 of HCMV) (48), apoptosis control (UL36 of HCMV) (45), and optimal virus replication in specific tissues (M43, M139, M140, and M141 of murine CMV) (22). This family includes a cluster of genes consisting of the HCMV ORFs UL23, UL24, UL28, and UL29 that are located at the left end of the betaherpesvirus genomes. Thus, UL26 not only shows homology to US22 proteins but is also located within a gene block of US22 family members. Reading frames with high similarity to UL26 could be detected within the genomes of murine (41), rat (52) and tupaia CMV (3), thus strengthening the assumption that the encoded proteins play an important role during viral replication.
Since no data on gene expression from the UL26 gene locus were available, we started our analysis by performing Northern blot experiments. This revealed a rather complex pattern of RNA expression: several transcripts that appear with an overall early-late kinetics during the HCMV replicative cycle could be detected. These transcripts could not be detected under the classical IE conditions of CHX treatment. All of these transcripts have the potential to code for pUL26 and could therefore contribute to the level of UL26 protein within an infected cell. Regulation of these transcripts was differential. One abundant transcript of 5.5 kb could be detected as early as 7 h after infection. Although this transcript was not detected under conditions of CHX block it could contribute to a rather early expression of UL26 during the viral replication cycle. Several true late RNAs were also observed with the UL26-specific probe. Remarkably, while the 2.65- and 0.75-kb RNAs showed an increase in abundance at late times after infection, expression of the 5.5-kb RNA was rather transient, with maximum RNA levels detected at 7 h after infection and a rapid decline at 24 h. A search for polyadenylation sequences 5′ and 3′ of the UL26 ORF revealed a sequence of ATTAAA ca. 200 nt downstream of the UL26 ORF; the only two other sequences conforming with a described polyadenylation signal were located upstream of the UL29 reading frame (see Fig. 4B). This suggests that the detected transcripts may have a coterminal 3′ end, as has previously been described for other transcription units of CMV (55, 58).
Prokaryotic expression of UL26 as a histidine-tagged protein revealed a polypeptide of 23 kDa, a finding which is in good agreement with the calculated molecular mass of 21 kDa. By Western blot analysis with a specific rabbit antiserum, two polypeptides of 21 and 27 kDa could be detected in cell lysates from HCMV-infected cells and cells that were transfected with a UL26 expression plasmid. Interestingly, there was a difference between infected cells and the isolated expression of UL26 in transfected cells. In transfected cells, the 27-kDa form of UL26 was more abundant than the 21-kDa form, whereas in infected cells the 21-kDa protein dominated. Initially, these results suggested to us that pUL26 is posttranslationally modified, most probably by phosphorylation. However, metabolic labeling experiments did not reveal phosphorylation of pUL26. Upon inspection of the sequence upstream of the predicted UL26 ORF, we noted an in-frame ATG that is located 34 amino acids 5′ of the predicted ATG. Since the 27-kDa form of pUL26 could no longer be detected after transfection or in vitro transcription-translation of a eukaryotic expression vector without the additional upstream ATG, we conclude that the two isoforms of pUL26 arise via alternative translation initiation at two in-frame start codons. Regulation at the level of translational initiation has been described for several cellular and viral genes (e.g., herpes simplex virus thymidine kinase gene) as a means to increase the diversity of protein products (20, 33, 37). Whether the two isoforms of pUL26 differ in function remains to be determined.
An association of pUL26 with viral particles has previously been noted by Baldick and Shenk (4). In that study, virion-associated proteins isolated from purified HCMV particles were used as substrates for chemical amino acid sequence analysis. A protein of 21 kDa was detected in preparations of dense bodies which are defective viral particles produced during high-multiplicity infection of fibroblasts and was subsequently identified as a gene product encoded by the UL26 ORF. By using the specific rabbit antiserum generated in our study, we could not only confirm the localization of pUL26 within dense bodies but could also demonstrate that both isoforms of pUL26 are constituents of infectious virus particles. Furthermore, we provide evidence that both isoforms of pUL26 are contained within the tegument of virus particles. There are two lines of evidence for this. First, the UL26 protein could not be solubilized by detergent treatment of virions as has been demonstrated for envelope proteins (31). Instead, it remained associated with the rather stable particles consisting of capsid and tegument proteins (18). Second, a comparison of virions and dense bodies showed that pUL26 is contained within both types of particles, whereas the major capsid protein is found only within virions (25, 31). While it is suggested that dense bodies do not have a full complement of tegument polypeptides, the important point here is that dense bodies are devoid of capsid proteins (25). This finally shows that pUL26 must be a constituent of the tegument of HCMV viral particles.
This study was initiated in order to identify HCMV-encoded proteins that are involved in the regulation of gene expression. Consistent with this, we observed that pUL26 acts as a transcriptional activator when tethered to the DNA-binding domain of the yeast factor GAL4. Transcriptional activation by pUL26 was detected by using an approach, termed activator trap, that is able to select strong transcriptional activation domains from a background of random DNA sequences (13, 19). The functionality of this screening strategy for the genomes of large DNA viruses was confirmed by the fact that we were able to isolate the previously known activation domain encoded by exon 3 of the IE1/2 gene region (38). However, the second described transcriptional activation domain of an HCMV-encoded protein, which is located at the carboxy terminus of IE2 (38), was not selected. This may be explained by the observation that the carboxy-terminal activation domain of IE2 is cryptic within the context of the intact protein requiring an exact fusion of a particular amino acid sequence to the GAL4 DNA-binding domain in order to be functional (38). This could have been missed by our screening assay. In contrast to similar selection strategies described for yeast, selection by the activator trap is based on mammalian cells (19, 34). This proved to be of particular importance for the identification of pUL26 since the UL26 activation domain is not active in yeast cells (K. Lorz and T. Stamminger, unpublished data), although our experiments classified UL26 as a domain with strong activity from a remote enhancer position in mammalian cells (12, 44). Furthermore, the UL26 activation domain appeared to be unusual since no sequences that are characteristic for known activation domains, such as clusters of serine and threonine, proline, glutamine, or acidic amino acids, could be detected. Thus, one might speculate that pUL26 interacts with a protein of mammalian cells that carries a conventional transcriptional activation domain, and it will therefore be of interest to identify the cellular interaction partners of this viral polypeptide.
Additional evidence for a role of pUL26 in modulating viral gene expression is provided by its nuclear localization and by the results of cotransfection experiments that detected an approximately 10-fold activation of the major IE enhancer-promoter in the presence of the UL26 protein. So far, we have not observed an activation of viral early or late promoters, suggesting that this effect is specific for the major IE enhancer-promoter. Since we found that pUL26 is present during the IE phase of viral gene expression due to protein import with the viral inoculum, this protein may play an important role during initial stages of viral replication. Whether pUL26 acts synergistically together with other tegument proteins of HCMV that are imported by viral infection, such as the pp71 (UL82) or ppUL69 transactivators, remains to be determined (32, 59).
In summary, we identified here a novel tegument protein of HCMV with the characteristics of a transcriptional activator that is imported during viral infection. To further determine the role of pUL26 for the regulation of HCMV replication, the construction of a recombinant virus with a deletion of the UL26 ORF is in progress.
Acknowledgments
We thank Donatella de Gaspero-Hoops for excellent technical assistance and B. Fleckenstein for continuous support.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB473) and the Bundesministerium für Forschung und Technologie (IZKF Erlangen). In addition, T.S. was supported by an EMBO short-term fellowship award.
REFERENCES
- 1.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 2.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr, U., and G. Darai. 2001. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J. Virol. 75:4854-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biegalke, B. J., and A. P. Geballe. 1991. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology 183:381-385. [DOI] [PubMed] [Google Scholar]
- 6.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 7.Britt, W. J., and L. Vugler. 1987. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J. Gen. Virol. 68:1897-1907. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Chiou, C. J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMarchi, J. M. 1981. Human cytomegalovirus DNA: restriction enzyme cleavage maps and map locations for immediate-early, early, and late RNAs. Virology 114:23-38. [DOI] [PubMed] [Google Scholar]
- 12.Escher, D., M. Bodmer-Glavas, A. Barberis, and W. Schaffner. 2000. Conservation of glutamine-rich transactivation function between yeast and humans. Mol. Cell. Biol. 20:2774-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escher, D., and W. Schaffner. 1996. Improved “activator trap” method for the isolation of transcriptional activation domains from random DNA fragments. BioTechniques 21:848-854. [DOI] [PubMed] [Google Scholar]
- 14.Fleckenstein, B., I. Müller, and J. Collins. 1982. Cloning of the complete human cytomegalovirus genome in cosmids. Gene 18:39-46. [DOI] [PubMed] [Google Scholar]
- 15.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerster, T., P. Matthias, M. Thali, J. Jiricny, and W. Schaffner. 1987. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 6:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazal, P., J. Young, E. Giulietti, C. DeMattei, J. Garcia, R. Gaynor, R. M. Stenberg, and J. A. Nelson. 1991. A discrete cis element in the human immunodeficiency virus long terminal repeat mediates synergistic trans activation by cytomegalovirus immediate-early proteins. J. Virol. 65:6735-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516-537. [DOI] [PubMed] [Google Scholar]
- 19.Gstaiger, M., and W. Schaffner. 1994. Strong transcriptional activators isolated from viral DNA by the ‘activator trap’, a novel selection system in mammalian cells. Nucleic Acids Res. 22:4031-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haarr, L., H. S. Marsden, C. M. Preston, J. R. Smiley, W. C. Summers, and W. P. Summers. 1985. Utilization of internal AUG codons for initiation of protein synthesis directed by mRNAs from normal and mutant genes encoding herpes simplex virus-specified thymidine kinase. J. Virol. 56:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagemeier, C., S. M. Walker, P. J. Sissons, and J. H. Sinclair. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385-2393. [DOI] [PubMed] [Google Scholar]
- 22.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermiston, T. W., C. L. Malone, P. R. Witte, and M. F. Stinski. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J. Virol. 61:3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 26.Irmiere, A., and W. Gibson. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J. Virol. 56:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 22:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner, R., H. Meyer, and M. Mach. 1989. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J. Virol. 63:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J., G. Prolla, A. Rostagno, R. Chiarle, H. Feiner, and G. Inghirami. 2000. Initiation of translation from a downstream in-frame AUG codon on BRCA1 can generate the novel isoform protein ΔBRCA1(17aa). Oncogene 19:2767-2773. [DOI] [PubMed] [Google Scholar]
- 34.Ma, J., and M. Ptashne. 1987. A new class of yeast transcriptional activators. Cell 51:113-119. [DOI] [PubMed] [Google Scholar]
- 35.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonough, S. H., and D. H. Spector. 1983. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology 125:31-46. [DOI] [PubMed] [Google Scholar]
- 37.O'Donovan, K. J., and J. M. Baraban. 1999. Major Egr3 isoforms are generated via alternate translation start sites and differ in their abilities to activate transcription. Mol. Cell. Biol. 19:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 40.Puchtler, E., and T. Stamminger. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1997. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolph, S. A., J. E. Kühn, K. Korn, R. W. Braun, and G. Jahn. 1990. Prokaryotic expression of the major capsid protein of human cytomegalovirus and antigenic cross-reactions with herpes simplex virus type 1. J. Gen. Virol. 71:2023-2031. [DOI] [PubMed] [Google Scholar]
- 43.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (“enhancer”) and proximal (“promoter”) positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spaete, R. R., R. M. Thayer, W. S. Probert, F. R. Masiarz, S. H. Chamberlain, L. Rasmussen, T. C. Merigan, and C. Pachl. 1988. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology 167:207-225. [DOI] [PubMed] [Google Scholar]
- 47.Stamminger, T., E. Puchtler, and B. Fleckenstein. 1991. Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J. Virol. 65:2273-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Triezenberg, S. J. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5:190-196. [DOI] [PubMed] [Google Scholar]
- 52.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wathen, M. W., and M. F. Stinski. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J. Virol. 41:462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wathen, M. W., D. R. Thomsen, and M. F. Stinski. 1981. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J. Virol. 38:446-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch, A. R., L. M. McNally, and W. Gibson. 1991. Cytomegalovirus assembly protein nested gene family: four 3′-coterminal transcripts encode four in-frame, overlapping proteins. J. Virol. 65:4091-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westin, G., T. Gerster, M. M. Muller, G. Schaffner, and W. Schaffner. 1987. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 15:6787-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler, M., T. aus dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkler, M., S. Schmolke, B. Plachter, and T. Stamminger. 1995. UL69 of HCMV, a homolog of the HSV ICP27, is contained within the tegument of virions and activates the major IE enhancer of HCMV in synergy with the tegument protein pp71. Scand. J. Infect. Dis. Suppl. 99:8-9. [Google Scholar]
- 60.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]