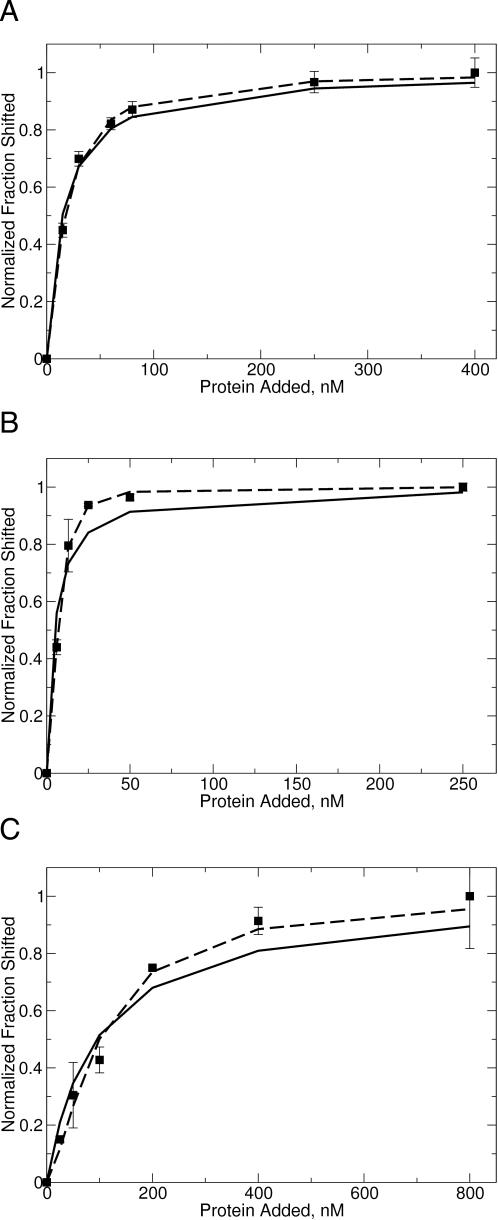
Figure 1.
Fitting of (A) unmethylated, (B) hemimethylated and (C) dimethylated DNA gel-shift data to various binding models. Data were fit as either M.RsrI monomer binding to DNA (solid line) or sequential binding of M.RsrI monomers (dashed line). The DNA used in the gel-shift experiments was a duplex 25 bp oligodexoxyribonucleotide with a single centered GAATTC target site bearing the specified methylation state at a concentration of 5 nM (17).