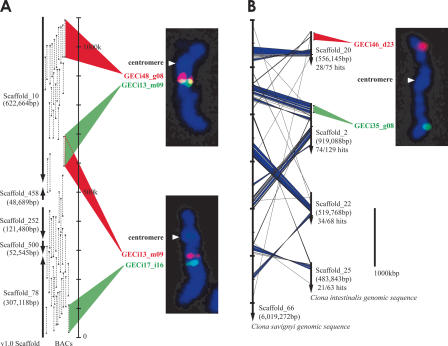
Figure 1.
Strategy for mapping BAC clones to chromosomes. (A) Correspondence between the distribution of BAC end sequences (BESs) on joined scaffolds and the cytogenetic location of FISH signals. The construction of joined scaffolds was performed using paired BAC end sequences. The left arrows indicate joined scaffolds from version 1.0 of the C. intestinalis draft genome. The regions of paired BAC end sequences corresponding to each scaffold are indicated by black triangles and are connected by dashed lines. For example, Scaffold_10 and Scaffold_458 are joined by two paired BAC end sequences in a tail-to-tail arrangement. Two-color FISH was performed using GECi48_g08 (red) and GECi13_m09 (green) BACs. The distribution of BESs on joined scaffolds indicates a gap of about 300 kb between the two clones, which gives an indication of the resolution achieved by FISH. GECi13_m09 (red) and GECi17_i16 (green) were also found to hybridize to the same chromosome. The cytogenetic locations of FISH signals correlate well to a first approximation with the order of joined scaffolds (Scaffold_10, Scaffold_458, Scaffold_252, Scaffold_500, and Scaffold_78). (B) A sample result from comparison of C. intestinalis and C. savignyi sequences. The vertical arrows represent scaffolds of the draft genomes (left) C. savignyi; (right) C. intestinalis. Diagonal lines connect best hit regions as determined by a FASTA comparison of C. intestinalis and C. savignyi sequences, with blue areas indicating regions with five sequential hits (within a distance of less than or equal to one gene), indicating strong similarity. Twenty-eight of 75 predicted genes in Scaffold_20 of C. intestinalis found best hit matches in Scaffold_66 of C. savignyi. Scaffold_2, Scaffold_22, and Scaffold_25 of C. intestinalis showed similar results, suggesting that Scaffold_20, Scaffold_25, Scaffold_22, and Scaffold_2 of C. intestinalis map to the same chromosome. These data corroborate results from comparative genomics studies that suggest a low rate of chromosome translocation between these two species. GECi46_d23 (red), which corresponds to Scaffold_20 and GECi35_g08 (green), which corresponds to Scaffold_2, map to different arms of the same chromosome.