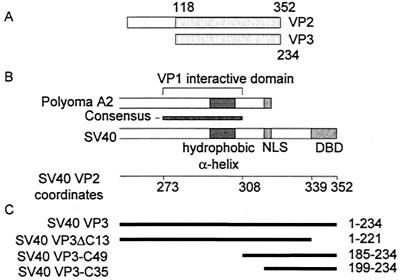
FIG. 1.
Schematic representation of VP2 and VP3. (A) Linear alignment of SV40 VP2 and VP3, showing the extent of a common segment and the N-terminal VP2-unique region. The numbers refer to amino acids of VP2 and VP3, respectively. (B) Alignment of the carboxy parts of polyomavirus A2 and SV40 VP2/3. SV40 amino acid coordinates (Swiss-Prot accession number P03093) are shown below the map. The consensus region (SV40 amino acids 273 to 308) is based on an alignment of eight polyomaviruses. The hydrophobic alpha-helix is almost identical in the polyomaviruses (4). The VP1 interactive domain of polyomavirus VP2/3 was determined by biochemical analysis (2) and X-ray crystallography (4). Abbreviations: NLS, nuclear localization signal (14); DBD, SV40 DNA-binding domain (amino acids 339 to 352) (5). (C) Schematic representation of VP3 truncation mutants. The heavy lines, aligned according to the schematic map in panel B, represent the amino acids included in the mutant proteins. The amino acids are designated at the right.