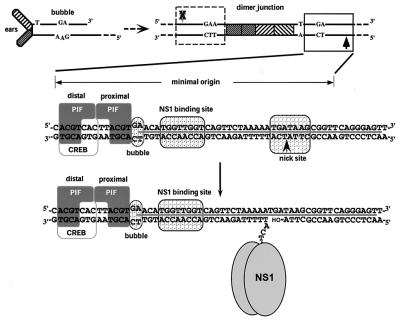
FIG. 1.
Formation and organization of MVM oriL. In the upper left is the structure of the left-end hairpin (oriLH) showing the 3′ hydroxyl group used for priming replication and the mismatched bubble sequence as present in the parental single-stranded viral genome. In the upper right is the organization of the left-end hairpin sequences within the duplex dimer junction generated by replication through the hairpin. The hatched boxes represent the palindromic sequences that were originally folded to give the ears of the hairpin form. The boxed sequence (expanded below) represents the minimum active replication origin on the outboard arm, oriLTC, while the sequence in the dashed box represents the corresponding origin on the inboard arm, oriLGAA, which is inactive. The arrows denote the potential nick sites on each side of the junction, solid for active and X-ed out shaded for inactive. The middle diagram illustrates the sequence of oriLTC, showing the different elements involved in replication. The PIF binding site (see the text) overlaps a consensus binding site for the CREB/ATF family of host transcription factors. The two tetranucleotide motifs bound by PIF are labeled “distal” and “proximal” to indicate their positions relative to the NS1 binding site. The other boxes indicate sequences involved in the bubble dinucleotide (or trinucleotide) spacer element, the ACCA2 NS1 binding site, and the nick site, a specific sequence required for nicking and covalent attachment of NS1. The heavy line between the DNA strands indicates sequences protected by NS1 from DNase I digestion. The position of the nick reflects the new determination described in this report. The bottom diagram shows the nicking and covalent attachment of an oligomer of NS1 via a phosphotyrosine bond. The nicking reaction liberates a 3′ hydroxyl group, which primes DNA synthesis mediated by host cell DNA polymerases.