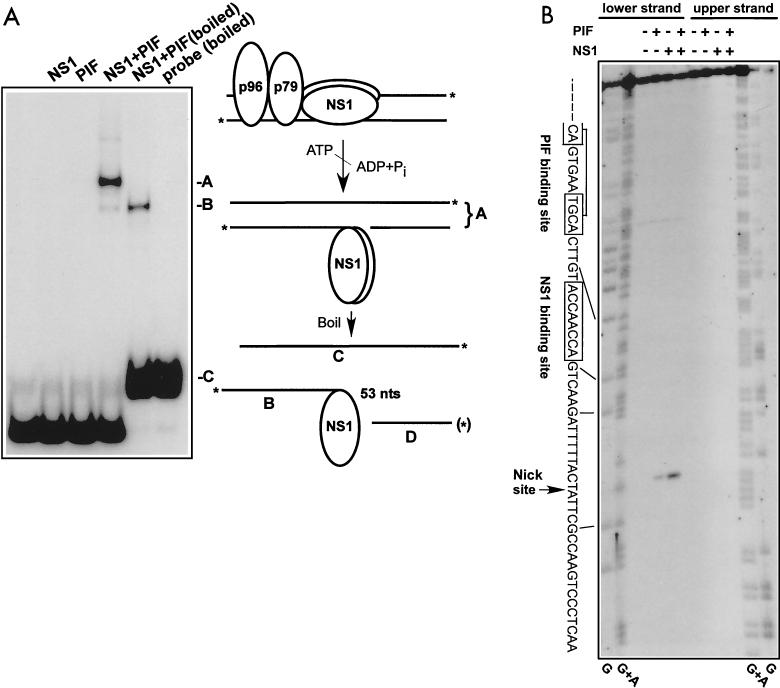
FIG. 3.
Characterization of the nick site in the minimal left-end origin. (A) A DNA fragment containing the oriLTC sequence was 32P labeled at its 3′ ends as described in Materials and Methods; incubated in the absence or presence of NS1 (100 ng), PIF (25 ng), and ATP (3 mM); and analyzed on a neutral SDS-polyacrylamide gel. Only covalent NS1-DNA complexes are significantly retarded in this gel system. For lane NS1+PIF (boiled), the nicking reaction mixture was denatured by boiling immediately before electrophoresis. A schematic model of the nicking reaction is depicted to the right of the gel autoradiograph. The asterisks indicate the positions of the 32P label at the 3′ end of each strand of the origin DNA. The letters A, B, and C correlate the DNA or DNA-protein structures produced in the reaction with bands on the gel. The fragment D labeled with an asterisk in parentheses indicates the 5′-end-labeled fragment analyzed in panel B. (B) DNA fragments containing the oriLTC sequence, separately labeled at the 5′ end of either the upper or lower strand (as depicted in panel A), were incubated with 3 mM ATP in the presence (+) or absence (−) of NS1 (100 ng) and PIF (25 ng) as indicated at the top of the gel. The samples were digested with proteinase K before analysis on a 6% denaturing polyacrylamide gel. Lanes G and G+A contain the products of G- and GA-specific chemical sequencing reactions of each labeled substrate and were used for aligning the position of the nick site. The sequence of the lower strand, depicted in panel A, is shown on the left side of the gel, and the lines indicate the position of this sequence in relation to the chemical sequencing reaction. An arrow indicates the putative nick site, representing the position of migration of fragment D in panel A. The boxed sequences show the positions of the specific NS1 and PIF binding sites.