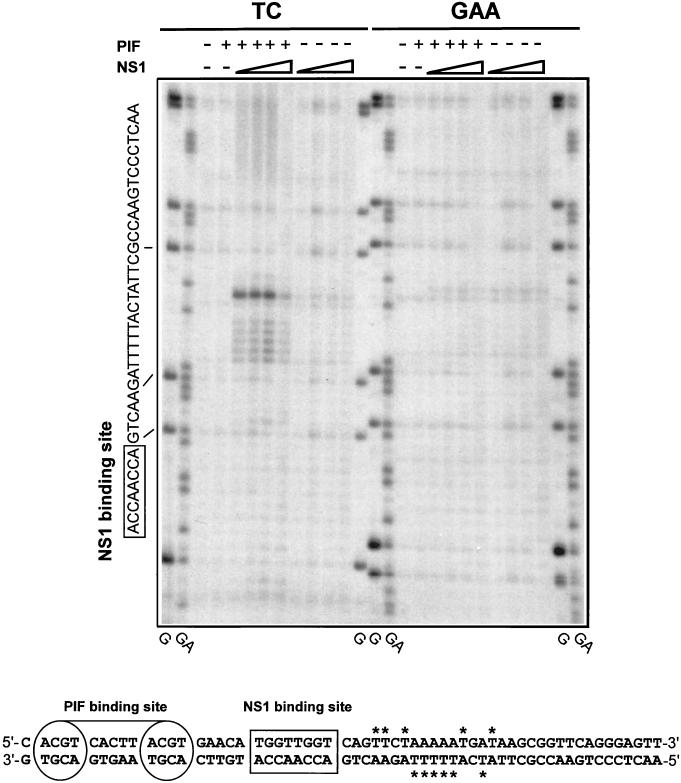
FIG. 4.
PIF and NS1 nicking of the minimal left-end origin causes local unwinding. DNA fragments containing either the oriLTC or oriLGAA sequence were 32P labeled at the 3′ end of the lower strand (Fig. 1) and incubated with 3 mM ATP in the presence (+) or absence (−) of NS1 (increasing amounts of purified GST-tagged NS1 [25, 50, 100, and 225 ng]) and PIF (50 ng of purified PIF p79-p96 complex) as indicated at the top of the gel. The samples were exposed to KMnO4 and subjected to alkaline hydrolysis before analysis on a 6% denaturing sequencing polyacrylamide gel. TC and GAA indicate the form of oriL that was used as a substrate in the left and right sets of KMnO4 assays, respectively. Lanes G and GA contain the products of G- and GA-specific chemical sequencing reactions of each labeled substrate and were used to align the positions of KMnO4-sensitive residues. The sequence of the relevant strand of the origin is shown on the left, and the lines indicate the alignment of this sequence to the chemical sequencing reaction. The boxed sequence shows the position of the specific NS1 binding site. At the bottom is a summary of the positions of KMnO4-sensitive thymidine residues (labeled with asterisks) detected in both strands of the origin.