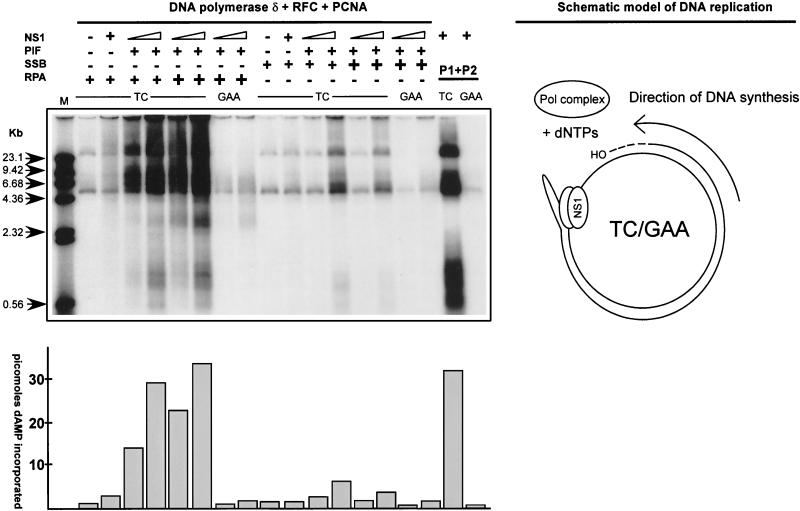
FIG. 7.
Reconstitution of replication initiated from the minimal left-end origin. Replication reaction mixtures were assembled using combinations of purified recombinant His-tagged NS1, PIF p79-p96 complex, RPA or E. coli SSB, RFC, PCNA Pol δ, or previously described phosphocellulose fractions of 293 cell extracts as positive controls (10) as indicated above the gel. Plasmid pL1-2 TC containing the minimal active left-end origin was the replication-positive template, and pL1-2 GAA was used as a negative control. The different purified proteins were titrated to achieve optimal replication. For PIF, RFC, PCNA, and Pol δ, constant amounts of 50, 125, 100, and 100 ng, respectively, were used in the assay. The increasing amounts of NS1, as indicated at the top of the gel, were 90 and 270 ng. RPA or E. coli SSB was added (+) in 0.5- or 1.5-μg amounts. Replicated DNA was digested sequentially with proteinase K and HindIII as described in Materials and Methods and then analyzed by agarose gel electrophoresis, along with the 32P-labeled λ phage DNA markers described in the legend to Fig. 6B. The amount of dAMP (in picomoles) incorporated into each replication product is indicated in the histogram at the bottom of the autoradiograph. A schematic model of the replication reaction is at the right.