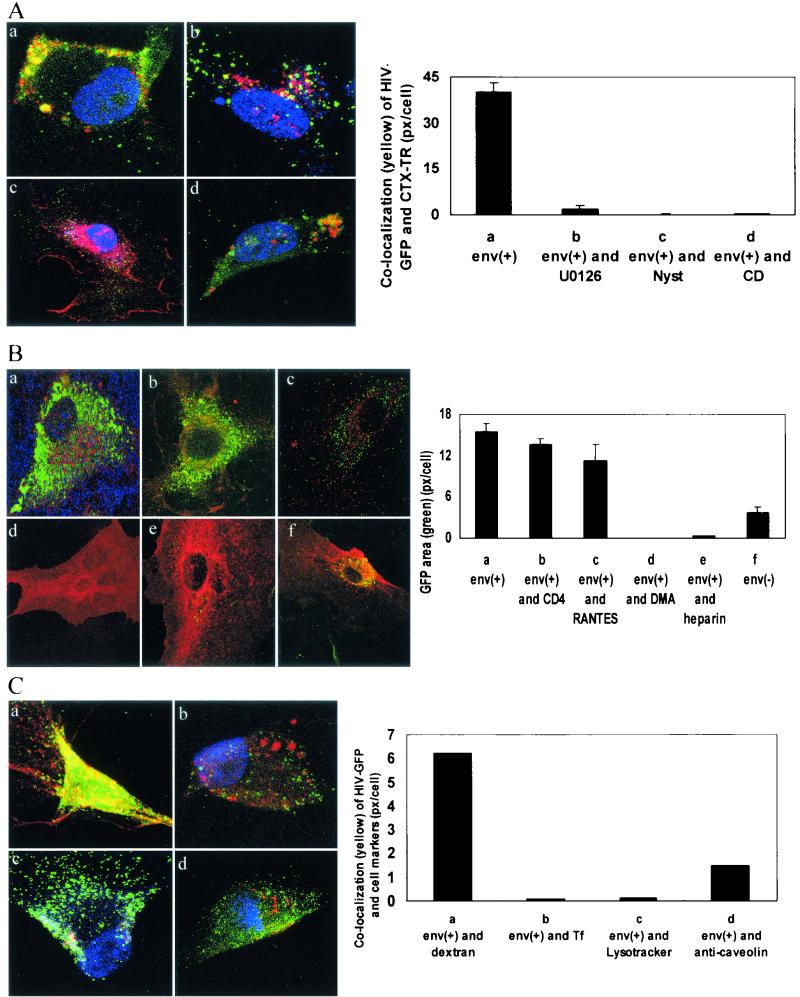
FIG. 8.
Virus entry by macropinocytosis. (A) HIV-1 Env+ binds to ganglioside GM1 and enters BMVECs by a process sensitive to inhibition by cholesterol-extracting agents or a MEK inhibitor. BMVECs were not pretreated (a) or were pretreated with U0126 (10 μM for 1 h) (b), nystatin (Nyst) (25 μg/ml for 1 h) (c), or cyclodextrin (CD) (10 mM for 1 h) (d). The cells were then infected with HIV-GFP Env+ (5 × 107 RNA copies); 2.5 h p.i., the cells were stained with cholera toxin B-TR (CTX-TR) (10 μg/ml) for 30 min and fixed, and three cells were then examined by confocal microscopy (magnification, ×95). Using an overlay of approximately 30 confocal sections from each examined cell, the area of colocalization (yellow) (pixels/cell) was scanned and is shown on the y axis of the graph. (B) HIV-1 Env+ binding and entry is not reduced by anti-CD4, is partially inhibited by AOP-RANTES, and is completely inhibited by heparin or dimethylamiloride. BMVECs either were not pretreated (a and f) or were pretreated for 30 min with anti-CD4 (10 μg/ml) (b), for 1 h with AOP-RANTES (100 nM) (c), or for 1 h with dimethylamiloride (DMA) (20 μM) (d). The cells were then infected with HIV-GFP Env+ (5 × 107 RNA copies) (a to d), HIV-GFP Env+ (5 × 107 RNA copies) preincubated for 5 min with heparin (20 μg/ml) (e), or HIV-GFP Env− (4.6 × 107 RNA copies) (f). Cholera toxin B-TR was added 2.5 h p.i., the cells were fixed 3 h p.i., three single sections were examined in each cell by confocal microscopy (magnification, ×95), and the area of GFP (green) (pixels/cell) was scanned and is shown on the y axis of the graph. (C) HIV-1 Env+ strongly colocalizes with fluid phase marker, weakly with caveolin and Lysotracker, and not at all with transferrin. The cells were infected with GFP-HIV-1 Env+; 2.5 h p.i., the cells were stained with TR-dextran (1 mg/ml) (a), TR-transferrin (20 μg/ml) (b), or Lysotracker dye (75 nM) (c), and anti-caveolin staining (d) was done after fixation. The cells were then examined by confocal microscopy (magnification, ×95); using an overlay of approximately 30 sections from each examined cell, the area of colocalization (yellow) (pixels/cell) was scanned and is shown on the y axis of the graph. The error bars indicate standard deviations.