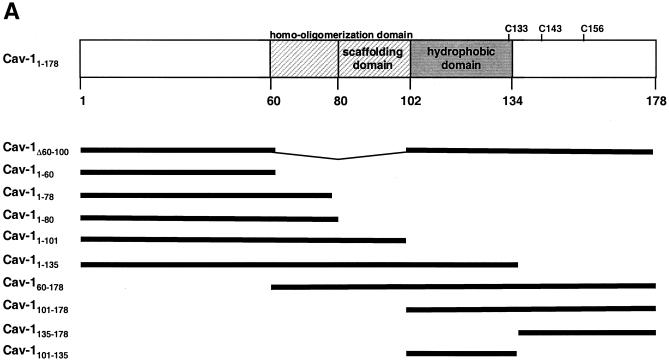
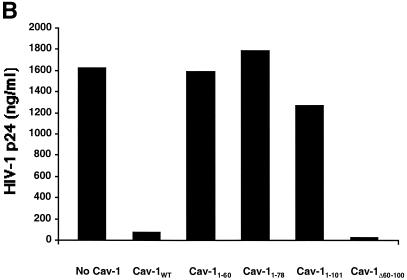
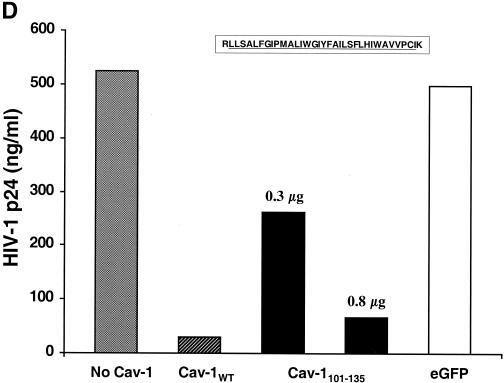
FIG. 6.
Mapping the Cav-1 effect by deletion mutagenesis. Shown are the structures of deletion mutants and effects of amino-terminal and hydrophobic domain deletions. (A) Structures of all Cav-1 deletion mutants used in the present study. Locations of palmitoylated cysteines mutated to alanine in the full-length protein are also shown (latter results described in text). (B) Cav-1 proteins lacking hydrophobic and carboxy-terminal regions do not inhibit HIV expression. (C) Amino-terminal Cav-1 residues proximal to the hydrophobic domain are dispensable. (D) The Cav-1 membrane-anchoring domain is necessary and sufficient to inhibit HIV expression. Residues 101 to 135 of Cav-1 are shown at the top of the panel, and the hydrophobic, putative membrane domain (residues 102 to 134) is underlined. As an additional control, cells in the experiment for panel D were cotransfected with 0.3 μg of pCMV.eGFP and 1.2 μg of pYU-2 (white bar). In panels B to D, equivalent Cav-1 mutant expression was verified by immunoblotting cell lysates with an anti-Myc MAb, and equal loading was verified by reprobing blots with an anti-α-tubulin antibody (data not shown). Unless otherwise indicated (D), 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV-myc (bars labeled “No Cav-1”), pCMV-myc.Cav-1 (bars labeled “Cav-1WT”), or pCMV-expressed Myc-tagged deletion-truncation mutants. p24 production was quantified in the cell supernatants at 40 h.