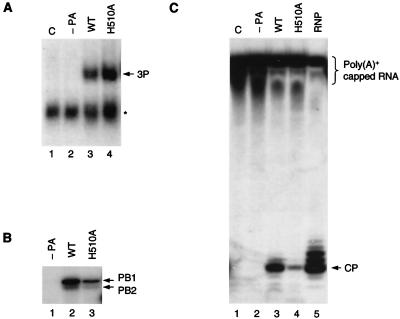
FIG. 5.
RNA-binding and endonuclease activities of the H510A polymerase. (A) 5′-end vRNA-binding activity of the H510A polymerase. Nuclear extracts from cells transfected either with pcDNA3 (lane 1), with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane 2), or with pcDNA-PB1, pcDNA-PB2, and either wild-type (WT) or H510A mutant pcDNA-PA (lanes 3 and 4, respectively) were used in gel shifts with 32P-labeled 15-nt 5′-end vRNA as described in Materials and Methods. The position of the polymerase complex (3P) is indicated. The star indicates a nonspecific band. (B) Capped RNA-binding activity of the H510A polymerase. His-tagged polymerases, partially purified from cells transfected either with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane1) or with pcDNA-PB1, pcDNA-PB2, and either pcDNA-PA-His6 wild-type (WT) or H510A mutant pcDNA-PA-His6 (lanes 2 and 3, respectively), were cross-linked to 32P-labeled capped RNA as described in Materials and Methods. The positions of the PB1 and PB2 bands are indicated. (C) Endonuclease activity of the H510A polymerase. Nuclear extracts from cells transfected either with pcDNA3 (lane 1), with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane 2), or with pcDNA-PB1, pcDNA-PB2, and either wild-type (WT) or H510A mutant pcDNA-PA (lanes 3 and 4, respectively) were incubated with poly(A)+-capped RNA, and cleavage products were analyzed as described in Materials and Methods. As a positive control, endonuclease cleavage was carried out with viral cores isolated from purified virus (RNP) (lane 5). The position of the specific cleavage product (CP) is indicated. The poly(A)+-capped RNA (indicated) runs as a smear due to the variety in the length of the poly(A) tail.