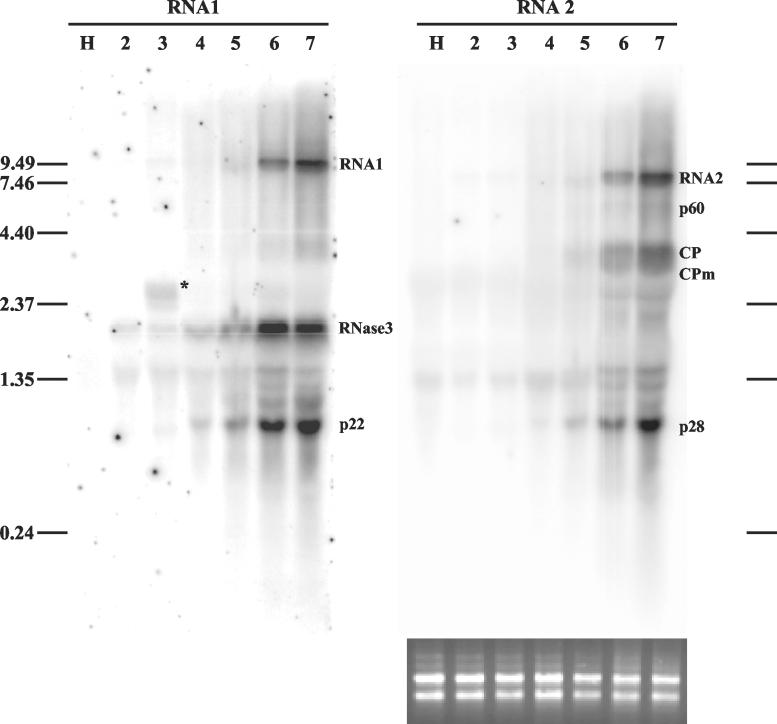
FIG. 2.
Northern analysis of genomic RNAs and sgRNAs of SPCSV in infected leaves of I. setosa. Total RNA extracted from SPCSV-infected leaves (lanes 2 to 7) and leaves of healthy plants (lane H) was first hybridized with an ssRNA probe corresponding to the negative strand of p28, the most 3′-proximal gene of RNA2. The same blot was subsequently used for hybridization with an ssRNA probe corresponding to the negative strand of p22, the most 3′-proximal gene of RNA1. Lanes 2 to 7 each contain total RNA extracted from six top leaves of the plant. The second leaf from the top (lane 2) was very young, was still folded, and had only very recently been invaded by SPCSV, in contrast to the lower leaves, which had been SPCSV infected for at least 2 weeks. The youngest leaf was too tiny for sampling. The positions of genomic RNA1 and RNA2 and the sgRNAs, identified by size, are indicated. The sgRNA corresponding to Hsp70h was not detected. The asterisk in lane 3 indicates a possible defective RNA (see text). The positions of the RNA molecular size markers (in kilobases) are indicated to the left and to the right. Equal loading of RNA was checked by ethidium bromide staining (shown below the RNA2 panel, the blot that was hybridized first).