Abstract
The African swine fever virus (ASFV) j4R protein is expressed late during the virus replication cycle and is present in both the nucleus and the cytoplasm of infected cells. By using the yeast two-hybrid system, direct binding, and coprecipitation from cells, we showed that the j4R protein binds to the alpha chain of nascent polypeptide-associated complex (αNAC). Confocal microscopy indicated that a proportion of j4R and αNAC interact in areas close to the plasma membrane, as well as through the cytoplasm in cells. In vitro binding studies suggested that binding of j4R to αNAC did not interfere with the binding of α- and βNAC subunits (the BTF3 transcription factor).
African swine fever virus (ASFV) is a large DNA virus which replicates in the cytoplasm of infected cells and contains a long, linear, double-stranded DNA genome encoding ca. 150 open reading frames (ORFs). The virus causes a hemorrhagic fever in domestic pigs but no disease signs in its natural hosts, warthogs, bushpigs, and soft ticks of the Ornithodoros species Ornithodoros moubata and O. erraticus, which act as vectors for the virus. ASFV has a replication strategy similar to that of poxviruses, but its structure is different, and it is classified as the only member of a new virus family, the Asfarviridae (9). ASFV encodes enzymes and factors needed for replication and transcription of the virus genome, as well as proteins that may play a role in interactions with the host that are important for virus survival and transmission (5, 40). Among the latter class of genes are those related to cellular genes, including genes encoding proteins that inhibit apoptosis, such as proteins related to IAP and Bcl2 (6, 26, 32). The ASFV A238L protein prevents activation of NF-κB-dependent gene transcription (29, 33, 37) and also binds to and inhibits calcineurin phosphatase activity. Calcineurin-dependent pathways, such as the activation of NFAT transcription factor, are therefore inhibited (21, 22). The ASFV EP402R protein resembles the host cell surface protein CD2 and is required for the adsorption of red blood cells around virus-infected cells and for virus-induced inhibition of bystander lymphocyte proliferation in response to mitogens (4, 5, 34).
The low level of amino acid similarity between ASFV-encoded proteins and other proteins makes predicting the function of virus-encoded proteins difficult. To facilitate this, we have identified host proteins that bind to virus-encoded proteins by using the yeast two-hybrid system.
The ASFV j4R protein has no significant homology with other proteins in the database, and to date, no functional data on the j4R protein have been published. In the present study we show that j4R is expressed late after infection and is conserved in the genomes of different virus isolates. We have also shown that the α chain of nascent polypeptide-associated complex (i.e., αNAC) binds to ASFV j4R protein.
The αNAC protein has roles in both translation and transcription. By binding to nascent polypeptide chains as they emerge from ribososmes, αNAC is proposed to prevent inappropriate targeting of polypeptides without signal sequences to the secretory pathway (38, 39, 43). αNAC has also been shown to act as a transcriptional coactivator potentiating transcription mediated by the c-Jun factor (23, 41). Here we demonstrate, by using recombinant proteins, that j4R binds directly to αNAC and, by coprecipitation, that j4R and αNAC are present in complexes in cells. Confocal microscopy suggested that a proportion of j4R and αNAC colocalizes in the cytoplasm in cells. By binding to αNAC the j4R protein may modulate either or both of these functions of αNAC.
MATERIALS AND METHODS
Cells and viruses.
ASFV isolates BA71V (11) and tissue culture-adapted Uganda (16) were used to infect either Vero or IBRS2 cell lines. The MVA T7 strain of vaccinia virus (36) was used to infect BSC40 cells. Field isolates of ASFV were used to infect porcine alveolar macrophages and were from Malawi LIL20/1, Bongera 83 (15, 35), Portugal Lisbon 57, Lisbon 60, Tomar 86 (gifts of J. D. Vigario, Laboratorio Nacional de Investigacao Veterinario, Lisbon, Portugal), Tanzania 87 (a gift of E. C. Anderson), South Africa RSA 85 (a gift of G. Thomson, VRI, Onderstepoort, Republic of South Africa), Mozambique 60, and Burundi 84. Cells were infected at a multiplicity of infection of three to five hemadsorption units per cell and were grown in Dulbecco modified Eagle medium (DMEM) containing HEPES in the presence of 10% fetal bovine serum.
Construction of plasmids.
The j4R ORF was amplified by PCR from a clone, LMw18, containing DNA from the ASFV Malawi LIL20/1 isolate (10) by using oligonucleotide primers (5′-GGGGGATCCATGGCCGGTCGTGTTA-3′ containing a BamHI restriction enzyme site [in boldface] and 5′-GGGGAATTCTATTTTTTTTCTCTATCA-3′ containing an EcoRI site [in boldface]). The PCR product was digested with EcoRI and BamHI and cloned downstream of the glutathione S-transferase (GST) gene in the pGEX2T vector (Amersham Pharmacia Biotech) to yield plasmid pGEX2Tj4R. Similarly, the j4R gene was cloned as a fusion with the Gal4 DNA-binding domain in the pGBT9 vector (Clontech) by using BamHI and EcoRI restriction sites included in the primers used for PCR to yield plasmid pGBT9j4R. The j4R gene was cloned downstream of the T7 promoter in the pcDNA3 vector (Invitrogen), also by using BamHI and EcoRI restriction enzyme sites, to yield pcDNAj4R. The porcine αNAC gene was amplified from pACT2 library clones isolated from a yeast two-hybrid screen with the oligonucleotide primers 5′-AAAGAATTCAAAGGATCCATGCCCGGCGAAGCCACA-3′ and 5′-AAACTCGAGTTACATTGTTAATTCCATAATCGC-3′. The PCR product was digested at EcoRI and XhoI sites included in the primers (in boldface) and cloned in frame with the T7 epitope tag in EcoRI and XhoI sites in the pET21A vector to yield plasmid pETαNAC. The EcoRI- and XhoI-digested PCR product was also cloned in the pcDNA3 vector digested with the same enzymes. A short double-stranded DNA (dsDNA) fragment formed by hybridization of complementary oligonucleotides and encoding the influenza virus hemagglutinin (HA) epitope tag was inserted upstream and in frame with αNAC at the KpnI and EcoRI sites to yield plasmid pcDNA HA αNAC. The porcine BTF3 gene was isolated from a yeast two-hybrid screen in the Gal4 activation domain vector pACT2. The gene was amplified by PCR and cloned in the pET34b vector (Novagen) by using EcoRI and HindIII sites included in the primers. Two versions of the genes BTF3a and -b were cloned that corresponded to the alternatively spliced variants (18) that are the same, except that the “b” form lacks sequences encoding the N-terminal 44 amino acids present in the 5′ end of the ORF. The primers used were 5′a (5′-GAGAATCGATGCGACGGACAGCGCA-3′) and 5′b (5′-GAGAATTCGATCAAAGAAACAATCATG-3′), and the same primer to the 3′ end of the ORF (5′-GAGAAGCTTAATTCAGTTTGCCTCATTCTT-3′) was used for each form of the gene.
Purification of recombinant GST-j4R and production of antibody.
Expression of recombinant GST-j4R fusion protein was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 to 4 h, and bacteria were lysed by sonication, followed by the addition of 1% Triton X-100. The fusion protein was purified by using glutathione-Sepharose beads and then eluted with 5 mM glutathione. New Zealand White rabbits were injected intramuscularly with 500 μg of GST-j4R fusion protein in Freund complete adjuvant and boosted after 3 weeks with 500 μg in Freund incomplete adjuvant. Rabbits were bled 1 and 3 months after the first inoculation. Immunoglobulin G was purified from whole serum on protein A-Sepharose columns.
In vitro binding assays.
The BL21 Lys E strain of Escherichia coli harboring either the αNAC gene cloned as a fusion with the T7 tag in the pET21A vector or the BTF3a or -b gene cloned as a fusion with the cellulose-binding domain (CBD) in the pET34b vector was grown at 37°C to an optical density at 600 nm of 0.4 to 0.6; 1 mM IPTG was added to induce recombinant protein expression, and incubation was continued for 2 h. Expression of the GST-j4R fusion protein from the pGEX2Tj4R plasmid in bacterial strain BL21 was induced similarly. Bacteria were harvested by centrifugation, extracts were prepared by freeze-thawing, and the insoluble fraction was removed by centrifugation. Concentrations of recombinant proteins were estimated by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels in comparison with known amounts of protein standards. Binding assays were performed by incubating 1 μg of T7-αNAC with anti-T7 antibody linked to agarose (Novagen) in phosphate-buffered saline (PBS) for 1 h. Beads were recovered by centrifugation and washed three times in the same buffer containing 0.1% NP-40, followed by a final wash in PBS. Various amounts of extracts containing GST-j4R or GST in PBS (0.5 to 10 μg) were added to beads, followed by incubation for 2 h at room temperature, and then were washed three times in PBS containing 0.1% NP-40. Beads were then boiled in SDS-PAGE sample buffer, and proteins were separated by SDS-PAGE and blotted onto a Hybond C membrane (Amersham Pharmacia). Alternatively, extracts containing 1 μg of CBD fusions with BTF3a or -b or CBD were incubated in PBS containing 0.1% NP-40 for 2 h at room temperature. The samples were then washed three times in the same buffer, boiled in SDS-PAGE sample buffer, separated by SDS-PAGE, and blotted onto a Hybond C membrane. In control experiments, GST-j4R or CBD-BTF3a or -b was bound directly to anti-T7 agarose beads without the addition of T7-αNAC. Recombinant proteins were identified by using anti-T7 monoclonal antibody (MAb) (Novagen) for the T7-αNAC protein, anti-GST (Sigma) for the GST-j4R protein, and anti-CBD MAb (Novagen) for CBD-BTF3a and -BTF3b fusion proteins and appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Dako), followed by detection by enhanced chemiluminescence (ECL).
Cell infections and transfections.
Cells were infected with virus at 3 to 5 PFU/cell for 1 h at 37°C and then incubated for various times in HEPES-modified DMEM containing 10% fetal bovine serum. Plasmid DNA (5 μg) was transfected into cells by using Lipofectin (Life Technologies) according to the manufacturer's recommendations.
SDS-PAGE and Western blotting.
Protein extracts were boiled in SDS-PAGE sample buffer (2% SDS, 10% glycerol, 62.5 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 0.01% bromophenol blue) and then separated on SDS-PAGE gels (10 or 12%). Gels were stained with Coomassie blue, soaked for 1 h in 1 M sodium salicylate, and dried down, and radioactive bands were then detected by fluorography. Alternatively, gels were transferred to a Hybond C membrane (Amersham Pharmacia Biotech); proteins were detected by reaction with primary antibody, followed by an appropriate HRP-conjugated secondary antibody, and then bound antibodies were detected by ECL.
Labeling of cells and immunoprecipitations.
For coprecipitation experiments, cells in six-well dishes were incubated in methionine- and cysteine-free medium for 20 min, labeled for 1 h by incubation in 0.83 MBq of [35S]Promix (530 MBq/ml; Amersham Pharmacia) per ml, and washed in PBS. Cells were then incubated in dithiobis(succinimidyl propionate) (DSP) cross-linking reagent (2.5 mM) for 1 h at room temperature and freeze-thawed at −70°C; Tris HCl (pH 7.5) was then added to a final concentration of 25 mM, and incubation was continued a further 30 min. NP-40 was added to a final concentration of 0.4%, and cell extracts were centrifuged at 13,000 rpm for 10 min to remove the insoluble material. Rat anti-HA MAb coupled to Sepharose (Roche) was added at a 1-in-40 dilution, and the mixture was incubated overnight at 4°C. Pellets were recovered by centrifugation and washed five times in 50 mM Tris (pH 7.5)-150 mM NaCl-5 mM EDTA-0.05% NP-40. Samples were boiled in SDS-PAGE sample buffer containing 5% β-mercaptoethanol and analyzed by SDS-PAGE.
Immunofluorescence.
For immunolabeling studies, cells were infected either with the BA71V strain of ASFV or with the MVA T7 strain of vaccinia virus and then transfected with plasmids pcDNA HA αNAC and pcDNAj4R singly or concurrently. At various times postinfection coverslips of cells were fixed in 3% paraformaldehyde for 20 min, quenched with 50 mM NH4Cl for 10 min, and permeabilized with 0.2% Triton X-100 in PBS for 5 min.
For immunolabeling, coverslips were incubated in 0.2% gelatin in PBS to block nonspecific binding and then incubated for 60 min in primary antibodies diluted in PBS-0.1% Tween 20. The antibodies used were rabbit anti-j4R (diluted 1:250) and rat anti-HA (diluted 1:2,000; Roche).
After three 5-min washes in PBS, bound antibody was detected with species-specific immunoglobulins conjugated to Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes/Cambridge Bioscience, Cambridge, United Kingdom), both of which were diluted 1:1,200 in PBS-0.2% Tween 20. Coverslips were again subjected to three 5-min washes in PBS, followed by 5 min in a 1:5,000 dilution of ToPro3 (Molecular Probes) in PBS and mounting in Vectashield (Vector Laboratories). Cells were imaged on a Leica TCS NT confocal microscope. Labeling controls included omission of primary antibody and adjustment of confocal imaging parameters to avoid all risk of cross talk between fluorophors.
Northern blotting of RNA from infected cells.
Cells were infected for 0, 4, 10, or 18 h, and total RNA was prepared by using extraction kits from Amersham Pharmacia Biotech. Poly(A) RNA was purified from total RNA by using oligo(dT) cellulose. mRNA (0.5 to 3 μg) was incubated at 65°C in 12.5 μl of deionized formamide, 2.5 μl of morpholinepropanesulfonic acid (MOPS) buffer (0.2 M MOPS [pH 7.0], 0.5 M sodium acetate, 0.01 M EDTA, 0.1% SDS), and 4 μl of 37% formaldehyde. The sample was chilled on ice and electrophoresed on a 1.2% agarose gel containing MOPS buffer and 2.2 M formaldehyde. RNA size markers were run in parallel and detected by ethidium bromide staining. Gels were blotted onto a Hybond N membrane (Amersham Pharmacia Biotech) in 0.4 M NaOH and hybridized with a probe prepared by random priming from a PCR fragment containing the j4R gene.
Yeast two-hybrid analysis.
The j4R gene and αNAC genes were cloned as fusions with the Gal4 DNA-binding domain in the pGBT9 vector (Clontech) and transformed into yeast strain Y190 with a pig macrophage cDNA library in the Gal4 activation domain vector pACT2 (21). Clones encoding proteins that interacted with the bait protein were selected by growth on medium lacking histidine in the presence of 50 mM aminotriazole. The clones from the library that expressed fusion proteins that interacted with the bait fusion protein were isolated from yeast colonies by transformation into E. coli HB101 and growth of colonies on minimal M9 medium. Isolated library plasmids were retransformed into yeast with the bait plasmid, as well as other control plasmids encoding irrelevant proteins (ASFV A238L) and double transformants screened for expression of Gal4-dependent β-galactosidase and growth on medium lacking histidine. Inserts in clones were sequenced by using an ALF express DNA sequencer and autoread sequencing kit (Amersham Pharmacia Biotech).
RESULTS
The j4R gene is expressed late during ASFV infection.
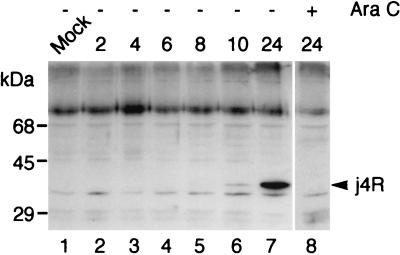
A rabbit antiserum was raised against the j4R protein to study the expression and localization of the protein during ASFV infection. The j4R gene was cloned as a fusion with the GST gene. Expression of the 67-kDa GST-j4R fusion protein was induced with IPTG, and the protein was purified by affinity chromatography by using glutathione-Sepharose and used to raise a polyclonal antiserum in rabbits. This antiserum reacted with both GST and the j4R protein cleaved from the GST-j4R fusion (data not shown). Extracts from cells harvested at various times postinfection with ASFV were separated by SDS-PAGE and probed by Western blotting with anti-j4R antiserum (Fig. 1). The j4R protein was detected as a protein of ca. 35 kDa in cell extracts harvested at late times postinfection (10 to 24 h). In extracts from cells incubated in the presence of cytosine arabinoside, an inhibitor of late gene expression, no j4R band was detected, thus confirming its late expression (Fig. 1).
FIG. 1.
The j4R protein is expressed late during ASFV infection. Extracts from IBRS2 cells either mock infected (Mock) or infected with the Uganda ASFV isolate for the indicated times (h) in the presence (+) or absence (−) of 50 μg of cytosine arabinoside/ml were separated by SDS-PAGE, and proteins were blotted to a Hybond C membrane. Blots were probed with anti-j4R serum (1 in 250 dilution), followed by goat anti-rabbit HRP-conjugated secondary antibody (1 in 2,000 dilution), and bound antibodies were detected by ECL. The positions of mass markers run in parallel are shown. The 35-kDa j4R protein is indicated by an arrow.
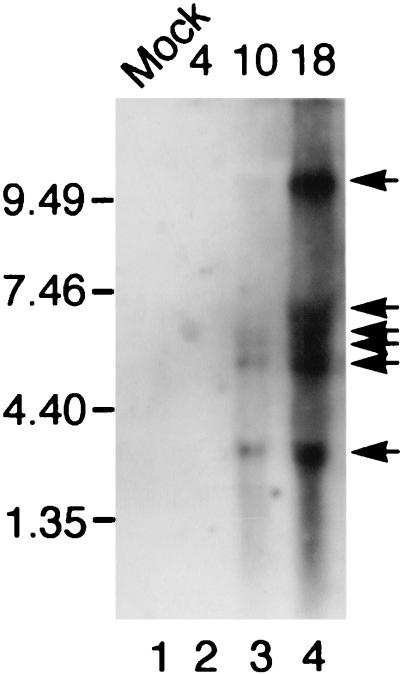
The j4R ORF is in the middle of a cluster of ORFs (j2R, j3R, j4R, j5R, j6R, and j7R) that are transcribed rightward. mRNA, extracted from cells at early and late times postinfection, was separated by agarose gel electrophoresis, blotted, and hybridized with a j4R gene probe (Fig. 2). There was no detectable hybridization of the probe with mRNA from uninfected cells and from cells harvested at 4 h postinfection. However, the probe hybridized to several discrete mRNA species, corresponding in size to about 3, 5, 6, 6.5, 7, and >10 kb, at late times postinfection. The signal for termination of ASFV gene transcription consists of a sequence of at least seven T's (1, 2). The various sizes of transcripts detected are therefore probably due to the termination of transcription, from the j4R and the upstream j2R and j3R gene promoters, at alternative sites. The nearest downstream signal for termination of transcription from j4R is on the noncoding strand of the ORF j8L, which is read in a leftward direction. Termination at this site would produce a transcript of ca. 3.4 kb, closest in size to the smallest transcript detected.
FIG. 2.
Northern blot of RNA from ASFV-infected cells hybridized with a j4R probe. Poly(A) RNA was extracted from IBRS2 cells that were either mock infected (Mock) or infected with ASFV Uganda isolate for the indicated time (4, 10, or 18 h). RNA was separated by agarose gel electrophoresis, blotted onto nitrocellulose, and hybridized with a fluorescein-labeled probe prepared by random priming from the j4R ORF. The positions of the size markers run in parallel are indicated. The positions of major bands are indicated by arrows.
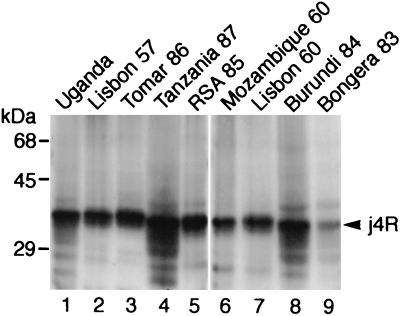
To determine whether the j4R protein is conserved in a number of ASFV isolates, pig macrophages were infected with nine virus strains isolated in different years between 1960 and 1986 and from six different countries in Europe and Africa. Extracts from these cells were separated by SDS-PAGE, blotted, and probed with anti-j4R serum (Fig. 3). A 35-kDa j4R protein was detected in extracts from cells infected with each of these isolates. The j4R gene from the virulent Malawi LIL20/1 isolate is the same length as the homologous gene from the Spanish tissue culture-adapted isolate BA71V, and the encoded proteins are closely related, sharing 96.2% amino acid identity (8, 40). These data indicate that the j4R gene is conserved in the genomes of different ASFV isolates.
FIG. 3.
Expression of j4R protein in cells infected with different ASFV isolates. Extracts from porcine alveolar macrophages that had been infected for 24 h with the ASFV isolates Uganda (lane 1), Lisbon 57 (lane 2), Tomar 86 (lane 3), Tanzania 87 (lane 4), RSA 85 (lane 5), Mozambique 60 (lane 6), Lisbon 60 (lane 7), Burundi 84 (lane 8), and Bongera 83 (lane 9) were separated by SDS-PAGE and blotted onto a Hybond C membrane. Blots were probed with anti-j4R serum (1 in 250 dilution) and then with goat anti-rabbit HRP-conjugated secondary antibody (1 in 2,000 dilution); bound antibodies were detected by ECL. The positions of molecular mass markers run in parallel are shown.
The j4R protein is present in both the nuclei and the cytoplasm of ASFV-infected cells.
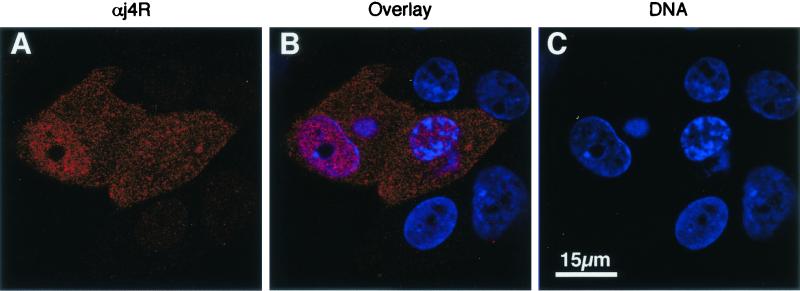
Antiserum raised against the j4R protein was used to determine the localization of the protein by immunofluoresence in ASFV-infected cells. Cells were infected and fixed with paraformaldehyde at 4, 6, 8, 12, 16, and 20 h postinfection. Permeabilized cells were stained with anti-j4R serum and Alexa Fluor 568-conjugated anti-rabbit serum. The j4R protein was detected in both the nuclei and cytoplasm of infected cells at late times (from 6 h) postinfection (Fig. 4). No significant change in this pattern of distribution was observed during this period.
FIG. 4.
Localization of j4R protein in ASFV-infected cells. Vero cells were infected with the BA71V isolate of ASFV, and at 12 h postinfection cells were fixed and labeled with anti-j4R antisera (red) and DNA with ToPro3 (blue) and imaged by confocal microscopy. Single optical sections are shown. Panels A and B show anti-j4R staining in red. Panels B and C show DNA in the nucleus and perinuclear virus factory areas stained with ToPro3 (blue). Panel B shows an image overlay.
Interaction of the j4R protein with host proteins.
To identify host proteins that bind to the j4R protein, the j4R gene was used as bait to screen a pig macrophage cDNA library by using the yeast two-hybrid system (21). The library clones encoding proteins interacting with j4R included 21 cDNAs encoding the αNAC (accession number X80909), three containing cDNAs with 89% nucleotide identity with the 14-3-3 epsilon chain (accession number U54778), and three containing cDNAs with 71% nucleotide identity with acid sphingomyelinase-like phosphodiesterase (accession number Y08135). Plasmids containing Gal4 activation domain fusions with each of these genes were isolated and retransformed into yeast, along with the j4R Gal4 DNA-binding domain plasmid and an irrelevant gene (ASFV A238L) fused to the Gal4 DNA-binding domain to confirm that the encoded proteins interacted specifically with j4R. The predicted sequence of the porcine αNAC protein was the same length, 215 amino acids, and differed at only two residues from sequences of the mouse and human αNAC proteins. Inserts in clones encoding αNAC varied in size between 600 and 1,200 bp. Apart from three clones, which did not encode the N-terminal 10, 12, or 33 amino acids, all encoded the complete αNAC protein but contained different lengths of 5′ and 3′ untranslated region. This showed that the clones had been independently isolated from the library.
j4R protein binds to αNAC protein in vitro.
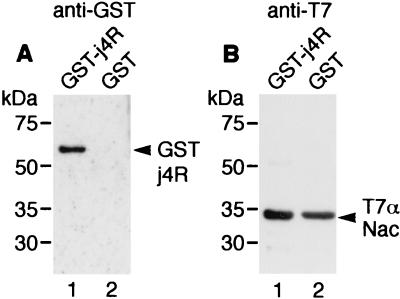
To confirm that j4R protein binds to αNAC directly and specifically, in vitro binding experiments were carried out. Recombinant αNAC protein fused to the T7 epitope tag in the pET21a vector was expressed in E. coli and purified by affinity chromatography by using anti-T7 MAb coupled to agarose beads. The T7-αNAC protein bound to anti-T7 antibody on agarose beads was mixed with bacterial extracts containing either GST-j4R or GST protein and washed in PBS containing 0.1% NP-40 to remove proteins not bound specifically to the T7-αNAC protein. Proteins bound to the column were eluted in SDS-PAGE sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting with anti-GST antibody and an anti-mouse HRP-conjugated secondary antibody, followed by ECL (Fig. 5A). Parallel blots were probed with anti-T7 antibody to confirm that the T7-αNAC protein had bound to the beads (Fig. 5B). Control experiments were carried out to confirm that GST-j4R did not bind to the anti-T7 agarose beads in the absence of the T7-αNAC protein (data not shown). This showed that αNAC binds to GST-j4R but not to GST, indicating that the interaction between j4R and αNAC is direct and specific. This interaction was stable in salt concentrations up to ca. 400 mM but was disrupted if detergent was added in the binding buffer (data not shown). However, if binding was carried out in the absence of detergent, the complexes could be washed in the presence of low concentrations of nonionic detergent (0.1% NP-40) without disrupting the interaction.
FIG. 5.
In vitro binding of j4R and αNAC. Recombinant T7-tagged αNAC protein was expressed in E. coli and purified by binding to anti-T7 agarose beads. Extracts from bacteria containing 0.5 μg of either GST-j4R fusion protein or GST were incubated with the T7-tagged αNAC on agarose beads (1 μg) for 1 h at room temperature in PBS and then were washed three times in the binding buffer containing 0.1% NP-40 and once in PBS. Bound proteins were eluted from the agarose beads with SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose Hybond C. (A) Blots that were probed with anti-GST antibody (1 in 4,000 dilution), followed by goat anti-mouse HRP-conjugated secondary antibody (1 in 200 dilution). (B) Blots probed with anti-T7 antibody (Novagen; 1 in 10,000 dilution), followed by goat anti-mouse HRP-conjugated secondary antibody (1 in 200 dilution) to detect the T7-tagged αNAC protein. Bound antibodies were detected by ECL. The positions of the molecular size markers are indicated.
j4R interacts with αNAC in cells.
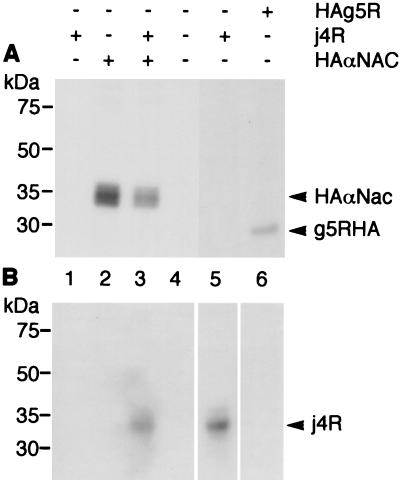
To investigate whether j4R is present in complexes with αNAC in cells, plasmids expressing an HA epitope-tagged αNAC gene and the j4R gene under control of the T7 promoter in the pcDNA3 vector were transfected into BS-C-1 cells, which had been infected with the MVA strain of vaccinia virus expressing T7 RNA polymerase (MVA T7) (36). The cells were labeled with [35S]methionine and [35S]cysteine and incubated with the thio-cleavable cross-linking reagent DSP to stabilize binding between interacting proteins in cells. Addition of cross-linking agent was necessary because nonionic detergent (0.4% NP-40) had to be included in cell lysis buffer to solubilize j4R protein, and this disrupts the interaction between j4R and αNAC. HA-tagged αNAC protein was precipitated with anti-HA MAb from cell extracts harvested 20 h after transfection. The cross-linking reagent was cleaved under reducing conditions, and immunoprecipitated proteins were separated by SDS-PAGE and detected by fluorography, or proteins coprecipitated with HA-tagged αNAC were detected, after blotting onto nitrocellulose, by probing them with anti-j4R serum (Fig. 6). The j4R protein was coprecipitated with HA-tagged αNAC from extracts from cells which had been transfected with both plasmids but not from cells transfected with either the j4R or HA-tagged αNAC expression plasmid alone. Control experiments were carried out in parallel with cells transfected with a plasmid expressing an irrelevant HA-tagged protein (the ASFV g5R protein [7]). The j4R protein was not coprecipitated with HA-g5R from these cell extracts (Fig. 6). These results indicate that j4R and αNAC are present in the same complex in cells. Without the addition of DSP, only small amounts of j4R were coprecipitated with αNAC (data not shown), probably because the nonionic detergent included in the cell lysis buffer disrupts binding between j4R and αNAC.
FIG. 6.
Coprecipitation of j4R and αNAC from transfected cells. Vero cells were infected with the MVA T7 strain of vaccinia virus and either transfected with plasmid pcDNA HA αNAC, pcDNAj4R, or pcDNA HAg5R or mock transfected. At 12 h after transfection cells were pulse-labeled with [35S]methionine-[35S]cysteine. (A) Fluorographs of proteins immunoprecipitated with anti-HA MAb (lanes 1 to 4 and 6; lane 5 contains extracts that had not been immunoprecipitated). (B) Parallel samples that were blotted onto nitrocellulose membranes and probed with anti-j4R serum, followed by goat anti-rabbit secondary antibody, after which bound antibodies were detected by ECL. The positions of molecular size markers run in parallel are shown.
Localization of j4R and αNAC in transfected cells.
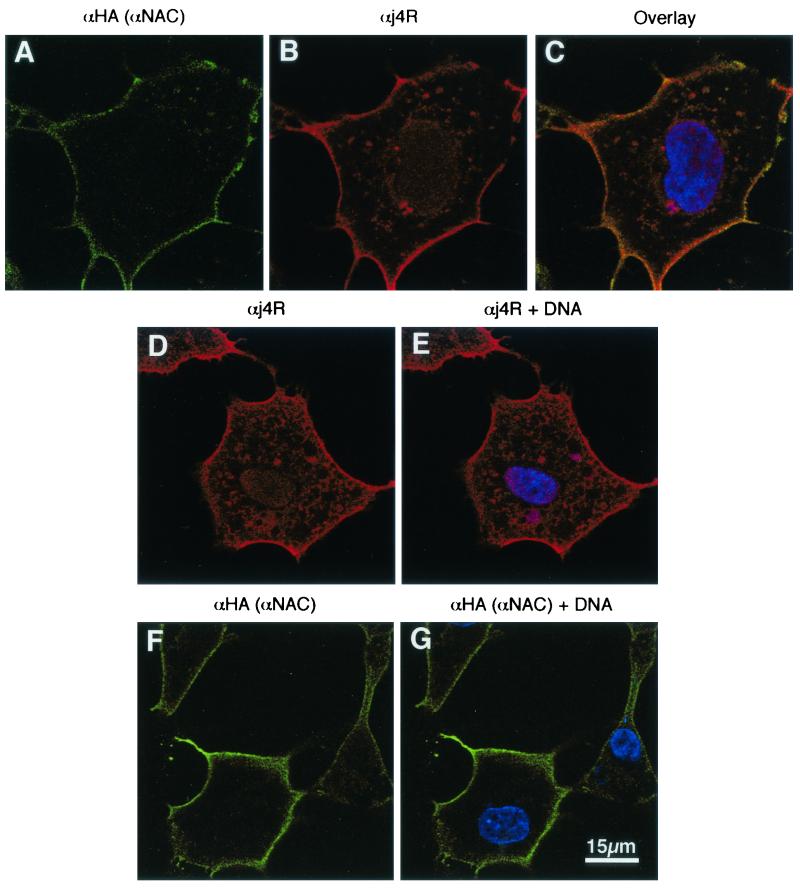
Plasmids expressing HA-tagged αNAC and j4R were transfected into MVA T7-infected cells, and at 12 h postinfection cells were fixed, permeabilized, and stained with anti-HA rat MAb and anti-j4R serum, followed by Alexa Fluor 488-conjugated anti-rat and Alexa Fluor 568-conjugated anti-rabbit immunoglobulin G. Fluorescent staining was visualized by confocal microscopy (Fig. 7). This showed that the distribution of HA-tagged αNAC was mainly cytoplasmic, with intense staining concentrated close to the plasma membrane. The j4R protein was more evenly distributed through the cell. In areas close to the plasma membrane, as well as other cytoplasmic regions, the j4R protein appeared to colocalize with the HA-tagged αNAC protein. These results indicate that a proportion of the expressed j4R and HA-tagged αNAC proteins colocalize in cells. The distribution of j4R and HA-tagged αNAC when expressed separately in cells was similar, suggesting that coexpression of j4R did not significantly alter the distribution of αNAC or vice versa (Fig. 7).
FIG. 7.
Localization of j4R and αNAC in cells. Cells were infected with the MVA T7 strain of vaccinia virus and transfected with plasmids pcDNA HA αNAC and pcDNAj4R together (A to C) or separately (D to G). At 12 h posttransfection cells were fixed and stained with anti-HA rat MAb to detect HA αNAC (green; panels A, F, and G) and anti-j4R serum (red; panels B, D, and E). DNA was stained with ToPro3 (blue; panels C, E, and G). Overlaid images of (i) panels A and B, plus DNA, are shown in panel C; (ii) panel D, plus DNA, are shown in panel E; and (iii) panel F plus DNA stain are shown in panel G. Cells were imaged by confocal microscopy, and single optical sections are shown.
Identification of host proteins that bind to αNAC by using the yeast two-hybrid system.
To help define possible functions of αNAC in porcine macrophages and hence predict the effect of j4R on αNAC function, we identified host proteins that bind to αNAC by using the yeast two-hybrid system to screen a porcine macrophage cDNA library (21). Clones encoding proteins binding to αNAC included 12 containing cDNAs with 96% nucleotide identity with the BTF3 transcription factor (accession numbers X53280 and X53281). This is present in a heterodimer with αNAC in cells (38, 43). In addition, three clones containing cDNAs with 82% nucleotide identity compared to IP63 protein (accession number X99330), one clone containing a cDNA with 90% nucleotide identity compared to ubiquitin hydrolase (accession number X98296), and one clone containing cDNA with 91% nucleotide identity compared to Ran BPM (accession number AB008515) were isolated from colonies encoding αNAC interacting proteins. The plasmids encoding αNAC interacting proteins were isolated and retransformed into yeast to confirm that the encoded proteins interacted specifically with αNAC and not other irrelevant proteins (ASFV A238L). Two differentially spliced forms of the BTF3 protein (a and b) are expressed in cells; the “b” form lacks the N-terminal 44 amino acids of the “a” form (18). Previously, each form of BTF3 has been shown to exist in a complex with αNAC (39). The IP63 protein is a leucine-rich protein of unknown function; ubiquitin hydrolases cleave ubiquitin from ubiquitin conjugates and Ran BPM is a Ran G protein-binding protein that is present in centrosomes. Expression of Ran GTP but not Ran GDP can induce microtubule self-organization (25, 27).
Binding of j4R to αNAC does not interfere with the formation of an αNAC, BTF3 complex.
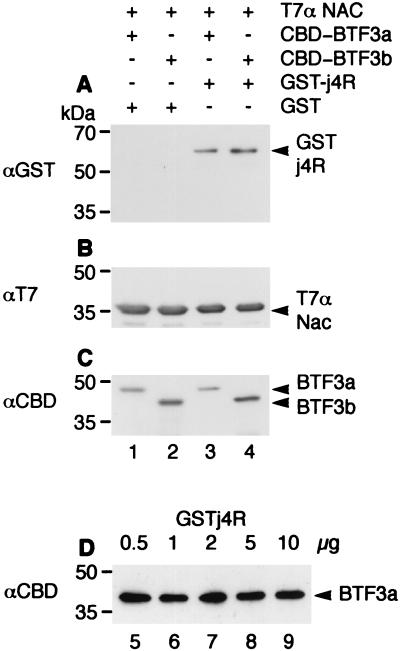
Fragments encoding full-length BTF3a and BTF3b were cloned in the pET34b vector as a fusion with a sequence encoding the CBD. The fusion proteins were expressed in E. coli and used in in vitro binding studies to determine whether the j4R protein interferes with binding of BTF3a or -b to αNAC. The purified T7-tagged αNAC protein (0.5 μg) was mixed with recombinant GST-j4R protein (0.5 to 10 μg). Recombinant BTF3a or -b fused to the CBD was then added (0.5 μg). Complexes containing T7 αNAC were purified by using anti-T7 agarose beads and then washed in PBS buffer containing 0.1% NP-40, separated by SDS-PAGE, and blotted with antibodies that recognize T7- αNAC, CBD-BTF3 fusion protein, or GST-j4R protein (Fig. 8). The results confirmed that both CBD-BTF3a and -b proteins bind to αNAC (Fig. 8), whereas no binding was observed between the CBD protein alone and αNAC (data not shown). Also, no binding was observed between the CBD-BTF3a and -b proteins and the anti-T7 beads alone (data not shown). The amount of BTF3a or -b bound to αNAC was not decreased when j4R was first bound to αNAC (Fig. 8), even in experiments in which up to a 20-fold excess of GST-j4R was added compared to T7-αNAC and CBD-BTF3 (Fig. 8). These results suggest that j4R protein does not interfere with the binding of αNAC to BTF3 under these in vitro conditions.
FIG. 8.
Binding of j4R to αNAC does not inhibit binding of BTF3 to αNAC. Recombinant T7-αNAC protein was expressed in E. coli and purified by binding to anti-T7 agarose beads. Extracts from bacteria containing recombinant GST protein (0.5 μg) or GST-j4R protein (0.5 μg) were either held at 4°C (lanes 1 and 2) or mixed with T7-αNAC protein on agarose beads (0.5 μg) for 2 h at room temperature in PBS containing 0.1% NP-40 (lanes 3 and 4) and then washed three times in binding buffer and once in PBS. Extracts from bacteria containing recombinant CBD-BTF3a (0.5 μg) or CBD-BTF3b (0.5 μg) were incubated with the beads for 2 h at room temperature in PBS, washed three times in binding buffer, and then washed once in PBS. Proteins bound to the beads were eluted with SDS-PAGE sample buffer, separated by SDS-PAGE, and blotted onto a Hybond C membrane. Blots were probed with anti-GST (1 in 4,000 dilution), followed by goat anti-mouse HRP-conjugated antibody (1 in 200 dilution) (A); with anti-T7 antibody (Novagen; 1 in 10,000 dilution), followed by HRP-conjugated goat anti-mouse antibody (B); or with anti-CBD antibody (Novagen; 1 in 5,000 dilution), followed by HRP-conjugated goat anti-rabbit (1 in 2,500 dilution) (C). Bound antibodies were detected by ECL. In panel D, lanes 5 to 9, increasing amounts (0.5, 1, 2, 5, and 10 μg, respectively) of GST-j4R protein were added to T7-αNAC (0.5 μg) before the addition of CBD-BTF3a (0.5 μg). Proteins bound to T7-αNAC were collected on anti-T7 agarose beads; the beads were washed as described above, and bound proteins were separated by SDS-PAGE, blotted, and probed with anti-CBD antibody to detect bound BTF3a. The position of molecular size markers run in parallel is shown.
DISCUSSION
Our results show that the ASFV j4R protein is expressed late during infection and is present in both the nuclei and the cytoplasm of ASFV-infected cells. The protein is expressed by a wide range of different ASFV isolates and is conserved in length and sequence. Insights into the possible function of j4R were gained by identifying host proteins that bind j4R by using the yeast two-hybrid system. A large number of clones encoding j4R-interacting proteins contained cDNAs of the αNAC gene. NAC consists of α and β subunits and was first identified as a complex which binds to ribosomes and nascent polypeptides as they emerge from the ribosome. This interaction is important for correct targeting of polypeptides to the secretory pathway and to mitochondria (14, 19, 38). The β subunit of NAC binds to ribosomes, but both subunits are required to prevent cross-linking of the signal recognition particle to signal-less nascent polypeptide chains (3). A role for the βNAC homologue from Drosophila in translation has also been demonstrated (20). Interestingly, the NAC subunits also function as transcription factors. βNAC was first identified in HeLa cell extracts as a basal transcription factor, BTF3 (42). The αNAC subunit functions as a transcriptional coactivator potentiating c-Jun-mediated transcription in mammalian cells. αNAC is expressed in many adult tissues but specifically in bone cells during development (23). Expression of an alternatively spliced form of the αNAC gene, skNAC, is upregulated in differentiating myoblasts and osteoblasts and in cells involved in wound healing, and this has been shown to act as a transcriptional activator (24, 41, 42). The homologues of α- and βNAC in Saccharomyces cerevisiae also have roles in transcription and enhance DNA binding by the Gal4 transcriptional activator (17, 28, 30).
We confirmed that the interaction between j4R and αNAC is direct and specific by using purified recombinant proteins in in vitro binding assays. Evidence that the two proteins interact in cells was provided by coprecipitation of j4R and αNAC. These coprecipitation experiments and confocal microscopy indicated that only a proportion of both αNAC and j4R interact in cells. Most of the αNAC protein was present in the cytoplasm and was concentrated around the cell periphery, j4R was more evenly distributed through the cell and colocalized with αNAC in areas close to the plasma membrane, as well as other cytoplasmic locations. Previous reports have suggested that αNAC is mainly located in the cytoplasm, where it is present at concentrations of 3 to 10 μM (3). These studies have used biochemical fractionation of cells and in vitro binding to demonstrate an interaction between αNAC and ribososmes. However, only a proportion of αNAC was found associated with ribosomes. Two reports (3, 41) have used immunofluorescence to study the localization of αNAC but did not use confocal microscopy as in our study. Our experiments were carried out by transient expression of αNAC. Since αNAC is known to be present at high concentrations in cells (ca. 3 to 10 μM), transient expression is not likely to increase dramatically the intracellular concentration of αNAC and alter its distribution. However, we cannot exclude this possibility. The distribution of j4R in ASFV-infected cells (Fig. 4) compared with in transfected cells (Fig. 7) is slightly different since the relative proportion of j4R close to the plasma membrane is lower in the ASFV-infected cells. We do not have an explanation for this, although there are several possibilities. First, it may be because of different levels of expression of the j4R protein in infected compared to transfected cells, or second, the j4R protein may interact with another ASFV-encoded protein, resulting in a different distribution of j4R. Finally, ASFV infection may trigger signals within the cell, which results in a different distribution of j4R. Nuclear localization of αNAC has been reported after serum starvation of osteoblastic cells, suggesting that localization is cell cycle dependent and that αNAC localizes to the nucleus in cells arrested at the G0/G1 border (41). In yeast, single subunits of αNAC were transported to the nucleus when binding to ribosomes was inhibited (31). Thus, nuclear translocation of αNAC may be cell type, state, or signal dependent, and the signals regulating this translocation have yet to be fully defined. In our experiments we detected very little αNAC in the nucleus, and this distribution was not altered in the presence of j4R. Instead, most αNAC was localized close to the plasma membrane. The αNAC protein does not contain predicted transmembrane domains, and this localization is presumably due to interactions with other cellular proteins. There are several examples of transcription factors and other proteins involved in signaling pathways which are recruited to protein complexes close to the cytoplasmic tails of transmembrane receptors and following activation are translocated to the nucleus. The distribution that we report for αNAC is consistent with a similar translocation model for αNAC, although there are alternative possibilities.
Our results with in vitro binding assays suggested that binding of j4R to αNAC does not interfere with binding of BTF3 to αNAC. It may be that the binding of j4R to αNAC prevents interaction of αNAC with other proteins important for its function. In studies documenting the role of αNAC as a transcriptional coactivator, interactions between αNAC and both c-Jun transcription factor and TATA box-binding protein were reported (41). It is possible that binding of j4R may interfere with the binding of αNAC to these proteins. In our two-hybrid screen, as well as interactions between αNAC and its β subunit, BTF3, we identified interactions between αNAC and the IP63 protein, a ubiquitin hydrolase and a Ran G protein-binding protein. We have not investigated these interactions further, although they may provide further clues as to how αNAC and j4R function.
The functional significance of the interaction between j4R protein and αNAC remains unclear, and j4R may modulate either or both of the functions proposed for αNAC. However, it is more difficult to envisage an advantage for the virus in modulating the specificity of targeting of nascent polypeptides to the secretory pathway. In contrast, modulating the ability of αNAC to act as a transcriptional coactivator may enable the virus to inhibit transcriptional activation of host genes. c-Jun is involved in the transcriptional activation of many immunomodulatory genes (12). ASFV encodes one protein, A238L, which both inhibits activation of the NF-κB host transcription factor and, by inhibiting calcineurin phosphatase activity, also inhibits activation of NFAT transcription factor (21, 29). Since the primary target cells for ASFV replication in vivo are macrophages, which play a key role in activating host response to infection, virus interference with activation of host macrophage gene transcription may potentially have a profound effect on macrophage function.
Acknowledgments
We thank P. Wilkinson and G. Hutchings for ASFV isolates, Charles Abrams for helpful discussions, and Steven Archibald for help with figures.
Support for this study was provided by the BBSRC, the DEFRA, and the EU (QLK3-2000-00362).
REFERENCES
- 1.Almazán, F., J. M. Rodríguez, G. Andrés, R. Pérez, E. Viñuela, and J. F. Rodriguez. 1992. Transcriptional analysis of multigene family 110 of African swine fever virus. J. Virol. 66:6655-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. Rodríguez, A. Angulo, E. Viñuela, and J. F. Rodriguez. 1993. Transcriptional mapping of a late gene coding for the p12 attachment protein of African swine fever virus. J. Virol. 67:553-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatrix, B., H. Sakai, and M. Wiedmann. 2000. The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275:37838-37845. [DOI] [PubMed] [Google Scholar]
- 4.Borca, M. V., G. F. Kutish, C. L. Afonso, P. Irusta, C. Carrillo, A. Brun, M. Sussman, and D. L. Rock. 1994. An African swine fever gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption. Virology 199:463-468. [DOI] [PubMed] [Google Scholar]
- 5.Borca, M. V., C. Carrillo, L. Zsak, W. W. Laegreid, G. F. Kutish, J. G. Neilan, T. G. Burrage, and D. L. Rock. 1998. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J. Virol. 72:2881-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun, A., F. Rodríguez, J. M. Escribano, and C. Alonso. 1998. Functionality and cell anchorage dependence of the African swine fever virus gene A179L, a viral bcl-2 homolog, in insect cells. J. Virol. 72:10227-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartwright, J. L., S. T. Safrany, L. K. Dixon, E. Darzynkiewicz, J. Stepinski, R. Burke, and A. G. McLennan. 2002. The g5R (D250) gene of African swine fever virus encodes a Nudix hydrolase that preferentially degrades diphosphoinositol polyphosphates. J. Virol. 76:1415-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, L. K., S. R. F. Twigg, S. A. Baylis, S. Vydelingum, C. Bristow, J. M. Hammond, and G. L. Smith. 1994. Nucleotide sequence of a 55-kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL 20/1). J. Gen. Virol. 75:1655-1684. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Vinuela, and P. J. Wilkinson. 2000. Family Asfarviridae, p. 159-165. In M. H. V. van Regenmortel, C. M. Fauquet, et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom.
- 10.Dixon, L. K. 1988. Molecular cloning and restriction enzyme mapping of an African swine fever virus isolate from Malawi. J. Gen. Virol. 69:1683-1694. [DOI] [PubMed] [Google Scholar]
- 11.Enjuanes, L., A. L. Carrascosa, and E. Vinuela. 1976. Isolation and properties of the DNA of ASFV. J. Gen. Virol. 32:479-492. [DOI] [PubMed] [Google Scholar]
- 12.Foletta, V. C., D. H. Segal, and D. R. Cohen. 1998. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 63:139-152. [DOI] [PubMed] [Google Scholar]
- 13.Funfschilling, U., and S. Rospert. 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George, R., T. Beddoe, K. Landl, and T. Lithgow. 1998. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. USA 95:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haresnape, J. M. 1984. African swine fever in Malawi. Trop. Anim. Health Prod. 16:123-125. [DOI] [PubMed] [Google Scholar]
- 16.Hess, W. R., B. F. Cox, W. P. Heuschele, and S. S. Stone. 1965. Propagation and modification of ASFV in cell cultures. Am. J. Vet. Res. 26:141-146. [PubMed] [Google Scholar]
- 17.Hu, G. Z., and H. Ronne. 1994. Yeast BTF3 protein is encoded by duplicate genes and inhibits the expression of some genes in vivo. Nucleic Acids Res. 22:2740-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanno, M., C. Chalut, and J. M. Egly. 1992. Genomic structure of the putative BTF3 transcription factor. Gene 117:219-228. [DOI] [PubMed] [Google Scholar]
- 19.Lauring, B., H. Sakai, G. Kreibich, and M. Wiedmann. 1995. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92:5411-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markesich, D. C., K. M. Gajewski, M. E. Nazimiec, and K. Beckingham. 2001. bicaudal encodes the Drosophila βNAC homolog, a component of the ribosomal translational machinery. Development 127:559-572. [DOI] [PubMed] [Google Scholar]
- 21.Miskin, J. E., C. C. Abrams, L. C. Goatley, and L. K. Dixon. 1998. A viral mechanism for inhibition of the cellular phosphatase calcineurin. Science 281:562-565. [DOI] [PubMed] [Google Scholar]
- 22.Miskin, J. E., C. C. Abrams, and L. K. Dixon. 2000. African swine fever virus protein A238L interacts with the cellular phosphatase calcineurin via a binding domain similar to that of NFAT. J. Virol. 74:9412-9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau, A., W. V. Yotov, F. H. Glorieux, and R. St-Arnaud. 1998. Bone-specific expression of the alpha chain of the nascent polypeptide-associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol. Cell. Biol. 18:1312-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munz, B., M. Wiedmann, H. Lochmuller, and S. Werner. 1999. Cloning of novel injury-regulated genes. Implications for an important role of the muscle-specific protein skNAC in muscle repair. J. Biol. Chem. 19:13305-13310. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, M., H. Masuda, J. Horii, K. Kuma, N. Yokoyama, T. Ohba, H. Nishitani, T. Niyata, M. Tanaka, and T. Nishimoto. 1998. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 143:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neilan, J. G., Z. Lu, G. F. Kutish, L. Zsak, T. G. Burrage, M. V. Borca, C. Carrillo, and D. L. Rock. 1997. Inhibition of apoptosis by the African swine fever Bcl-2 homologue: role of the Bh1 domain. Virology 230:252-264. [DOI] [PubMed] [Google Scholar]
- 27.Nishimoto, T. 1999. A new role of Ran GTPase. Biochem. Biophys. Res. Commun. 262:571-574. [DOI] [PubMed] [Google Scholar]
- 28.Parthun, M. R., D. A. Mangus, and J. A. Jaehning. 1992. The EGD1 product, a yeast homolog of human BTF3, may be involved in Gal4 DNA binding. Mol. Cell. Biol. 12:5683-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell, P. P., L. K. Dixon, and R. M. E. Parkhouse. 1996. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70:8527-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reimann, B., J. Bradsher, J. Franke, E. Hartmann, M. Wiedmann, S. Prehn, and B. Wiedmann. 1999. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 15:397-407. [DOI] [PubMed] [Google Scholar]
- 31.Reimann, F. J., E. Hartmann, M. Kohler, and B. Wiedmann. 2001. Evidence for a nuclear passage of nascent polypeptide associated complex subunits in yeast. J. Cell Sci. 114:2641-2648. [DOI] [PubMed] [Google Scholar]
- 32.Revilla, Y., A. Cebrian, E. Baixeras, C. A. Martinez, E. Vinuela, and M. L. Salas. 1997. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domain. Virology 228:400-404. [DOI] [PubMed] [Google Scholar]
- 33.Revilla, Y., M. Callejo, J. M. Rodriguez, E. Culebras, M. L. Nogal, M. L. Salas, E. Vinuela, and M. Fresno. 1998. Inhibition of nuclear factor κB activation by a virus-encoded IκB-like protein. J. Biol. Chem. 273:5405-5411. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez, J. M., R. J. Yáñez, F. Almazán, E. Viñuela, and J. F. Rodriguez. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J. Virol. 67:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumption, K. J., G. H. Hutchings, P. J. Wilkinson, and L. K. Dixon. 1990. Variable regions on the genome of Malawi isolates of African swine fever virus. J. Gen. Virol. 71:2331-2340. [DOI] [PubMed] [Google Scholar]
- 36.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Nonreplicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 37.Tait, S. W. G., E. B. Reid, D. R. Greaves, T. E. Wileman, and P. P. Powell. 2000. Mechanism of inactivation of NF-κB by a viral homologue of IκBα: signal-induced release of IκBα results in binding of the viral homologue to NF-κB. J. Biol. Chem. 275:34656-34664. [DOI] [PubMed] [Google Scholar]
- 38.Wiedmann, B., H. Sakai, T. A. Davis, and M. Wiedmann. 1994. A protein complex required for signal sequence-specific sorting and translocation. Nature 370:434-440. [DOI] [PubMed] [Google Scholar]
- 39.Wiedmann, B., and S. Prehn. 1999. The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett. 458:51-54. [DOI] [PubMed] [Google Scholar]
- 40.Yanez, F. J., J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Vinuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]
- 41.Yotov, W. V., A. Moreau, and R. St-Arnaud. 1998. The alpha chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol. Cell. Biol. 18:1303-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yotov, W. V., and R. St-Arnaud. 1996. Differential splicing-in of a proline-rich exon converts a NAC into a muscle-specific transcription factor. Genes Dev. 10:1763-1772. [DOI] [PubMed] [Google Scholar]
- 43.Zheng, X. M., D. Black, P. Chambon, and J. M. Egly. 1990. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature 344:556-559. [DOI] [PubMed] [Google Scholar]