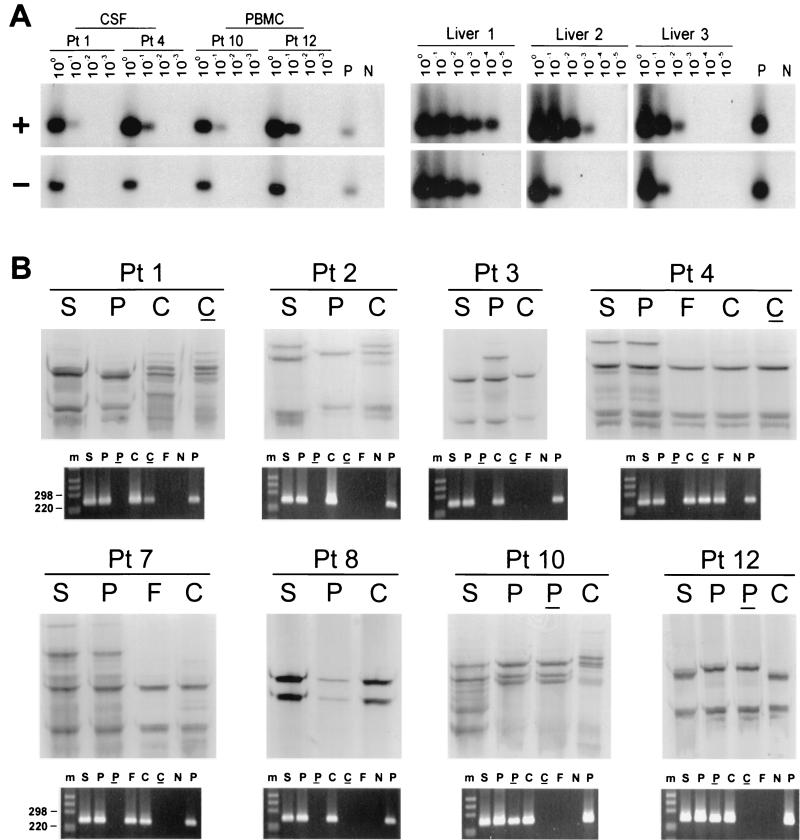
FIG. 1.
(A) Detection of HCV RNA positive strand (+) and negative strand (−) in CSF cells from patients (Pt) 1 and 4 and PBMC from patients 10 and 12, as well as in three liver tissue samples from HCV-infected patients. Tenfold serial dilutions of extracted RNA were tested for the presence of positive- and negative-strand HCV RNAs by Tth-based RT-PCR followed by Southern blotting and hybridization to a 32P-labeled probe. The amount of RNA loaded into the reaction mixture at dilution 100 corresponds to approximately 5 × 103 and 1 × 106 cells for CSF and PBMC samples, respectively. In the case of liver tissue samples, the first dilution corresponds to 1 μg of total RNA. Negative controls (lanes N) consisted of RNA extracted from PBMC and liver tissue from uninfected subjects, and positive controls (lanes P) consisted of 103 genomic eq of the correct synthetic strand mixed with 1 μg of RNA from an uninfected liver or RNA extracted from 106 PBMC. (B) Analysis by SSCP of 5′UTR HCV sequences amplified from serum (S), PBMC (P), and CSF from eight patients. Viral sequences were amplified from CSF cell pellets (C) for all patients and from CSF supernatant (F) for 2 patients. The RT-PCR products analyzed are also shown fractionated on NuSieve 2% agarose gels. Positive (P) and negative (N) controls consisted of RNA extracted from 106 PBMC from uninfected control subjects with and without the addition of 103 genomic eq of HCV RNA synthetic template. An underline represents the negative strand, and m represents the 1-kb molecular ladder (Gibco/BRL).