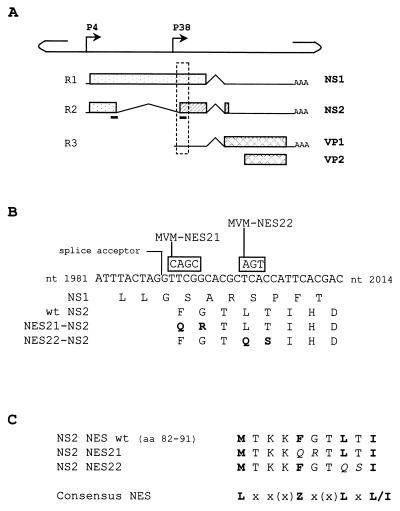
FIG. 1.
Mutagenesis strategy in the NES site of NS2. (A) The line scheme of the single-stranded MVMp genome shows the positions of the nonstructural gene promoter (P4) and the capsid gene promoter (P38). The three classes of RNA transcripts encoded by MVM, denoted R1 through R3, with introns (∧), polyadenylation sites (AAA), and open reading frames (boxes) encoding NS1, NS2, VP1, and VP2 proteins, are aligned below. The two black bars below the R2 transcript indicate the position of the NES site within the NS2 coding sequence. The stippled frame indicates the region that is shown in greater detail in panel B. (B) DNA sequence between MVMp nt 1981 and 2014 and amino acid sequence for this region of open reading frames 3 (for NS1) and 2 (for NS2). NS2 exon 2 initiates at the indicated large intron splice acceptor site. Mutations introduced into this sequence to create the two MVM-NES(−) genomes are shown in boxes above the wild-type sequence, and the resulting substitutions within the NS2 coding sequence are indicated in the last two lines, with the modified amino acids highlighted in bold. (C) Alignment of the wild-type and mutated NS2 NES sites with the NES consensus sequence (9). Leucine-like hydrophobic residues are shown in bold, and amino acid substitutions within the mutated NS2 NES sequences are shown in italics. x, any amino acid; Z, Met, Leu, Ile, Phe, or Val.