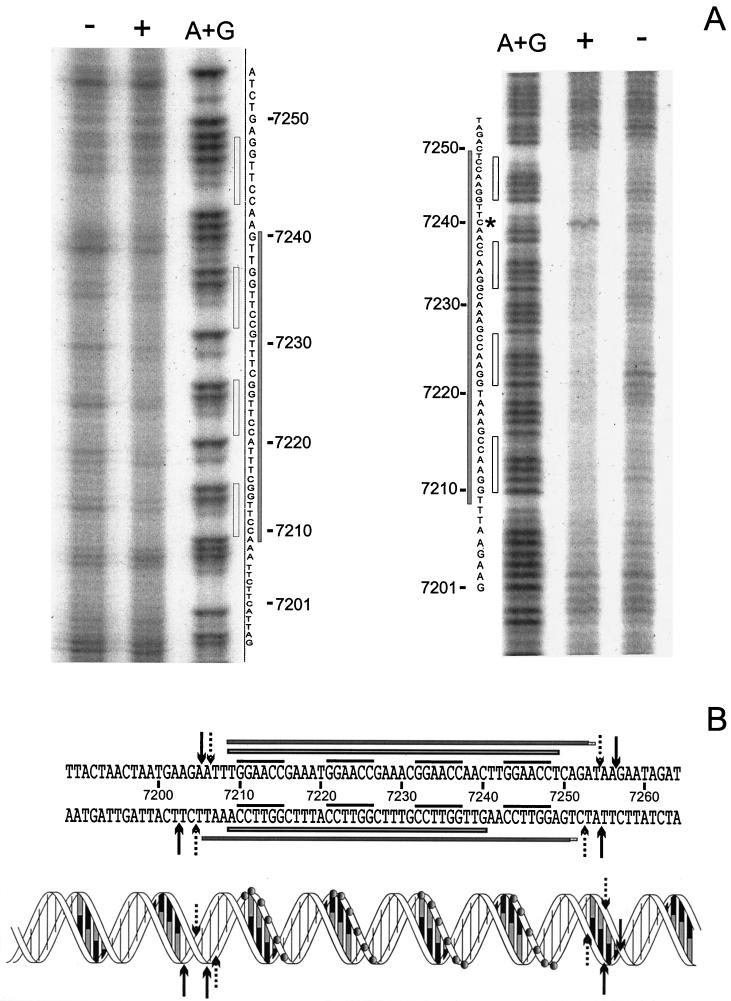
FIG. 6.
(A) Phenanthroline-copper footprinting analysis of the Pro11-orf11F5R2 complex. The left panel shows noncoding strand (inverted in order to facilitate the comparison); the right panel shows coding strand. + and −, presence or absence of Pro11, respectively; A+G, lane containing the products of an A+G Maxam and Gilbert sequence reaction; grey bars, protected regions; empty bars, position of hexamers; asterisk, hypersensitive residue C7240. (B) Summary of DNase I and phenanthroline-copper footprinting results. The sequence of orf11 from position 7189 to 7264 (base numbering is according to the numbering of the published r1t sequence [GenBank accession number U38906]) is shown above, while a schematic representation of the double helix is given below. In the latter, grey circles indicate the phosphates of the hexamers. An empty bar on each strand indicates nucleotides protected from phenanthroline-copper cleavage. Filled bars on each strand indicate the region protected from DNase I digestion. The four hexamers are indicated by the overlining. DNase I digestion sites important to define the boundaries of the protected regions are indicated by either dotted (protected sites) or black (unprotected sites) arrows.