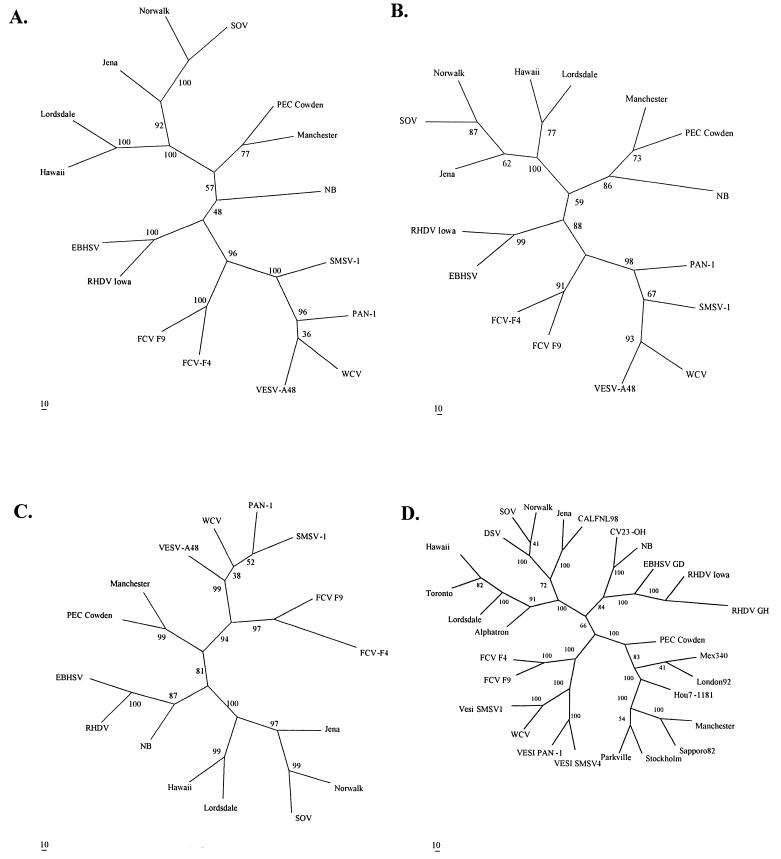
FIG. 2.
Unrooted consensus trees of 2C-like helicase/ATPase (A), 3C-like protease (B), 3D-like RdRp (C), and major capsid (VP-1) proteins (D) of BEC-NB. The lengths and ORF-1 locations of the amino acid sequences used to prepare the multiple alignments of the viruses used in the comparisons are shown in Table 1. The final trees depicted represent a consensus of trees for 100 bootstrapped data sets analyzed using the maximum-likelihood algorithm. The bootstrap values indicate the percentage of replicates supporting the branch points predicted in the consensus trees.