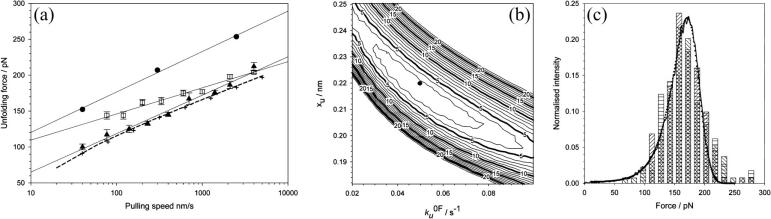
FIGURE 3.
(a) Speed dependence of the unfolding forces of (protein L)5 (▴), (I27*)5 (□), and (ubiquitin)9 (•). Error bars, where shown, represent ±SE of triplicate data sets. Solid lines through each data set are a best fit to guide the eye. Data for I27 and ubiquitin taken from Brockwell et al. (23) and Carrion-Vazquez et al. (3), respectively. Fitting the data for protein L to an analytical solution (dashed line, see Materials and Methods) estimates that the height and the position of the unfolding barrier relative to the native state is smaller and shorter (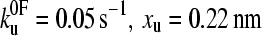 ) than that obtained for (I27*)5 (
) than that obtained for (I27*)5 (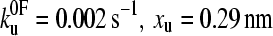 (23)). Monte Carlo simulations, using the best fit parameters for protein L obtained above, give identical modal values (cross-hairs) to those predicted by the analytical model. (b) Error analysis of parameter pairs reveals degeneracy in the fit of
(23)). Monte Carlo simulations, using the best fit parameters for protein L obtained above, give identical modal values (cross-hairs) to those predicted by the analytical model. (b) Error analysis of parameter pairs reveals degeneracy in the fit of 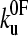 and xu to the observed experimental data for (protein L)5. Contour lines link parameter pairs calculated to have equal χ2 error. (c) The three experimental force frequency distributions at 1400 nm s−1 are consistent with those predicted by the analytical model (dotted lines) and Monte Carlo simulation (solid black line) using the parameter pair marked by a solid circle in b.
and xu to the observed experimental data for (protein L)5. Contour lines link parameter pairs calculated to have equal χ2 error. (c) The three experimental force frequency distributions at 1400 nm s−1 are consistent with those predicted by the analytical model (dotted lines) and Monte Carlo simulation (solid black line) using the parameter pair marked by a solid circle in b.