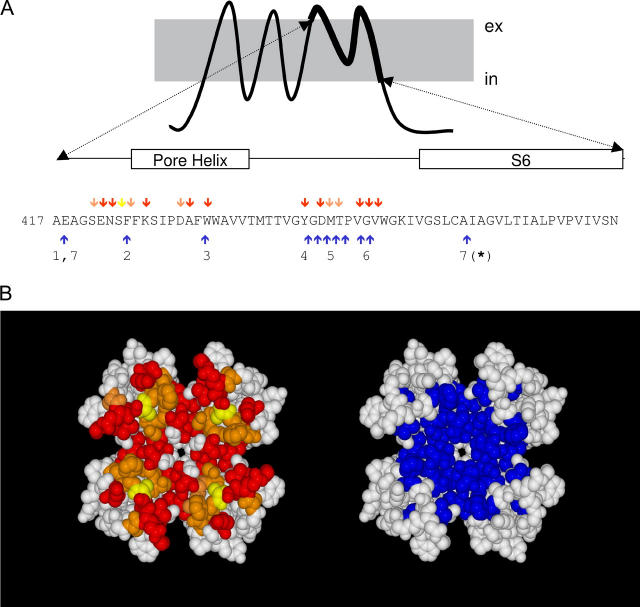
FIGURE 1.
Spatial overlapping between slow inactivation and peptide toxin binding. (A) Amino acid sequence of the H5 and S6 segments of Shaker K-channels. Red, yellow, and orange arrows point to residues important for slow toxin binding whereas blue arrows indicate residues important for slow inactivation. (B) Possible position of residues involved in slow inactivation (blue) and in peptide toxin binding (red, yellow, and orange) according to a homology modeling of the Shaker sequence on the KcsA structure template (www.expasy.org/spdv). Red residues have been shown to be important for the binding of α−KTx scorpion toxins (AgTx II and CTX). Yellow residues are important for κ−PVIIA binding, whereas orange residues are important for both types of toxins. Important peptide toxin binding residues are defined as those in which nonconservative mutations promote >8-fold changes in affinity. These data were taken from Goldstein et al. (27), Ranganathan et al. (28), and Gross and Mackinnon (47) for α-KTx and Scanlon et al. (29) and Jacobsen et al. (30) for κ−PVIIA. Residues important for slow inactivation were those in which conservative mutations promote >3-fold changes in the inactivation kinetics. Data for the slow inactivation were from (1) Ortega-Saenz et al. (49), (2) Perez-Cornejo (50), (3) Starkus et al. (7), (4) Heginbotham et al. (51), (5) Liu et al. (6), (6) Larsson and Elinder (48), and (7) Hoshi et al. (1) and Ogielska and Aldrich (52). Molecular rendition was by POVRAY 3.5 (www.povray.org).