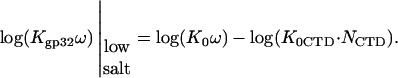TABLE 1.
Parameters describing the nonelectrostatic binding of gp32 and its mutants to single-stranded nucleic acids and to the gp32 C-terminal domain
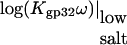 |
log(K0ω) | −log(K0CTDNCTD) | ΔG0CTD kcal/mol | |
|---|---|---|---|---|
| gp32/ss λ-DNA | 7.90 | 6.50 ± 0.20 | 1.4 ± 0.2 | 1.93 ± 0.20 |
| gp32/poly(A) | 7.15 | 5.75 ± 0.20 | 1.4 ± 0.2 | 1.93 ± 0.20 |
| K3A/poly(A) | 6.72 | 5.02 ± 0.20 | 1.7 ± 0.2 | 2.40 ± 0.20 |
| R4T/poly(A) | 6.10 | 4.10 ± 0.20 | 2.0 ± 0.2 | 2.60 ± 0.20 |