TABLE 1.
Summary of mean square displacement measurements, 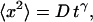 for the VirE2-ssDNA complexes under various treatments of the extracts
for the VirE2-ssDNA complexes under various treatments of the extracts
| Experiment No. | Substrate | Conditions | Prefactor | Exponent-γ | Movement |
|---|---|---|---|---|---|
| 1 | anVirE2 | Untreated | 0.59 | 0.99 | Active |
| 2 | anVirE2 | + AMP-PNP | 0.78 | 0.98 | Active |
| 3 | anVirE2 | + Cytochalasin D | 0.51 | 0.93 | Active |
| 4 | plVirE2 | + Cytochalasin D | 1.67 | 0.97 | Passive Brownian |
| 5 | plVirE2 | + Cytochalasin D + Nocodazole | 1.48 | 0.97 | Passive Brownian |
| 6 | plVirE2 | Untreated | 0.32 | 0.75 | Passive restricted |
| 7 | anVirE2 | + Nocodazole | 0.39 | 0.76 | Passive restricted |
| 8 | anVirE2 | + Vanadate | 0.26 | 0.76 | Passive restricted |
| 9 | anVirE2 | + Antidynein | 0.24 | 0.77 | Passive restricted |
Note that the measured exponents-γ cluster near either unity or 3/4. Active movement of the anVirE2 complexes (No. 1–3) always shows 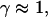 consistent with directed movement along a random network as explained in the text. Cases of thermally driven motion (No. 4,5) yield
consistent with directed movement along a random network as explained in the text. Cases of thermally driven motion (No. 4,5) yield 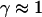 when F-actin has been depolymerized and
when F-actin has been depolymerized and 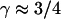 when F-actin is present (No. 6–9).
when F-actin is present (No. 6–9).
