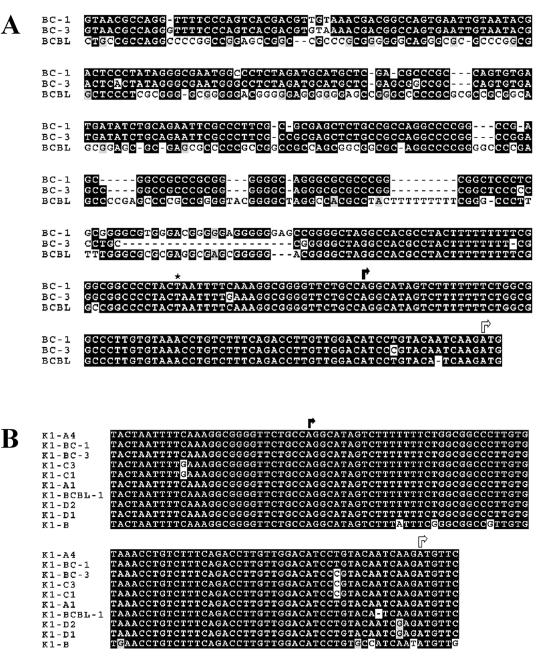
FIG. 1.
Characterization of the K1 promoter region in KSHV-positive BCBL-1, BC-1, and BC-3 cells. (A) A Clustal W alignment of sequences upstream of the K1 gene in BCBL-1, BC-1, and BC-3 cells is shown. Identical nucleotides are highlighted in black boxes, and divergent nucleotides are highlighted in gray boxes. Based on data from the 5′-RACE experiments, the transcription start site (black arrow) for K1 was assigned to a position 75 bp upstream of the translation start site (white arrow). The putative TATA box (asterisk) for transcription of K1 is located 25 bp upstream of the transcription start site. (B) GenBank sequences upstream of K1 from all four clades of KSHV isolates were aligned with the BCBL-1, BC-1, and BC-3 sequences by using the Clustal W alignment program. The identical nucleotides are highlighted in black boxes, and divergent nucleotides are highlighted in gray boxes. The transcription start site is depicted with a black arrow, and the translation start site is depicted with a white arrow.