Abstract
Fluoroquinolones target two bacterial type II topoisomerases, DNA gyrase and topoisomerase IV. Acquired resistance to quinolones occurs stepwise, with the first mutation occurring in the more sensitive target enzyme. To limit the emergence of resistance, quinolones should ideally possess dual activities against the two enzymes. For reasons that are as yet unclear, Staphylococcus aureus gyrase is less sensitive to quinolones than topoisomerase IV, counter to its greater sensitivity in Escherichia coli, thereby limiting the use of quinolones for the treatment of staphylococcal infections. Mutations in the α4-helix domain of the GyrA subunit of gyrase are important in determining quinolone resistance. We replaced an extended region encompassing the α4 domain in the E. coli GyrA protein with its homolog in S. aureus and tested for its ability to complement a thermosensitive gyrase and its catalytic and noncatalytic properties. Purified gyrase reconstituted with chimeric GyrA was more resistant to ciprofloxacin than wild-type gyrase at both inhibition of catalytic activity and stimulation of cleavage complexes, and this difference was more apparent in the presence of K+-glutamate. The chimeric GyrA subunit was able to complement thermosensitive gyrase, similar to wild-type GyrA. Without supplemental K+-glutamate the MICs of ciprofloxacin for thermosensitive E. coli complemented with chimeric DNA gyrase were equal to those for E. coli complemented with wild-type gyrase but were twofold higher in the presence of K+-glutamate. Our findings suggest that the extended α4 domain of S. aureus GyrA is responsible, at least in part, for the increased resistance of S. aureus gyrase to quinolones and that this effect is modulated by K+-glutamate.
The roles of DNA gyrase and topoisomerase IV in quinolone action and resistance can be assessed in cells by selection of resistant mutants with exposure to a quinolone. In Escherichia coli, first-step resistance mutations map in the genes encoding gyrase subunits (7, 30), usually gyrA, implying that gyrase is the primary drug target in cells and is more sensitive than topoisomerase IV to fluoroquinolone attack. Nalidixic and oxolinic acids were found to inhibit the supercoiling activity of purified gyrase from wild-type cells but not from resistant gyrA mutants (7, 30). These gyrA mutations were sufficient alone to cause a substantial increase in quinolone resistance (100-fold for nalidixic acid and 10- to 30-fold for ciprofloxacin). In contrast, in Staphylococcus aureus, parC or parE mutants selected with many quinolones are sufficient alone to cause resistance in a gyrA+ background (4, 5, 22, 32), and gyrA mutations alone are silent (22), contributing incrementally to fluoroquinolone resistance only in the presence of parC or parE mutations. Catalytic assays with purified enzymes corroborate the findings of studies with whole cells that have shown that in gram-negative bacteria topoisomerase IV is less sensitive than gyrase to many quinolones (3, 16, 24). Although the sensitivities of the topoisomerase IV enzymes of E. coli and S. aureus (by use of the same DNA substrate) to many fluoroquinolones are similar, S. aureus gyrase appears to have substantially reduced intrinsic sensitivity to quinolones, reversing the relative sensitivities of the two enzymes and explaining why topoisomerase IV is the primary fluoroquinolone target in S. aureus (3).
Quinolone resistance occurs stepwise by mutations in the two topoisomerase target enzymes, with the first mutation generally occurring in the more sensitive enzyme (12). If the original sensitivities of both DNA gyrase and topoisomerase IV were the same, no single mutational alteration in either enzyme would result in an increase in the MIC (11, 22); rather, resistance would require concurrent alterations in both enzymes. Because double mutations are a rare genetic event (occurring at a frequency of 10−14 to 10−16 for fluoroquinolones), the preferential use of fluoroquinolones with dual activities could limit the selection of fluoroquinolone resistance in wild-type bacteria (27, 29, 37). There are, however, few fluoroquinolones that possess such a dual targeting property for gram-positive bacteria (14, 23), and the apparent low sensitivity of S. aureus gyrase to most fluoroquinolones deters the efficient use of current members of the quinolone class against this pathogen.
The biochemical basis for the lower sensitivity of S. aureus gyrase to quinolones in comparison to that of E. coli gyrase is not yet known. It could lie in factors involved in the DNA-enzyme-quinolone ternary complex, as well as in the cellular milieu in which these reactions take place or a combination of multiple factors. The hot spot for quinolone resistance mutations in type II topoisomerases is the α4 domain of the GyrA and ParC subunits. There are a number of differences in the α4 domains of E. coli and S. aureus GyrA subunits. Studies with purified E. coli gyrase indicate that mutations in this domain reduce drug binding (1, 34, 36). Swapping of the same α4-helix domain of E. coli ParC with that of E. coli GyrA has been shown to decrease the catalytic efficiency of topoisomerase IV for decatenation but also to confer increased sensitivity to topoisomerase IV-mediated cleavage complex formation in the presence of a quinolone, norfloxacin, suggesting that this domain can affect quinolone action (25). Therefore, we have examined if the difference in the α4-helix domain of gyrase between E. coli and S. aureus is responsible for the reduced sensitivity of the latter to quinolones. In this work we replaced an extended α4 domain in E. coli gyrase with its homolog in S. aureus and tested for the ability of the chimeric protein to complement a thermosensitive gyrase and for its catalytic and noncatalytic properties. Our findings suggest that the extended α4 domain of S. aureus GyrA is responsible, at least in part, for the increased quinolone resistance of S. aureus gyrase and that this effect is modulated by K+-glutamate, an amino acid that is present in higher concentrations in S. aureus cells than in E. coli cells.
MATERIALS AND METHODS
Bacterial strains, vectors, and growth conditions.
Escherichia coli strains were grown in Luria-Bertani medium (Beckton Dickinson, Sparks, MD). All strains except E. coli KNK453 were grown at 37°C. E. coli KNK453, which encodes a thermosensitive GyrA subunit ([nalA43 (G751D)(Ts) F− polA thyA uvrA phx]) (18) was grown at 30°C and 43°C, as indicated. Ampicillin was used at 100 μg/ml. Minimal medium contained 47.8 mM Na2HPO4 · 7H2O, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 4 mg/ml glucose, and 1 μg/ml thiamine. Casamino Acids (10 mg/ml; Difco, Detroit, MI) were added as supplements, as indicated. Thymidine (150 μM) was added to support the growth of KNK453 cells.
Complementation assay and drug susceptibility determinations.
pBAD24, an arabinose-inducible expression vector (8) with the individual gyrB and gyrA genes cloned into it, was transformed into E. coli KNK453 and propagated at 30°C. For the complementation assays, 10-fold dilutions of an overnight culture were serially inoculated on minimal medium plates supplemented with thymidine, and the plates were incubated at 43°C. Ciprofloxacin was kindly provided by Bayer Corporation (West Haven, CT). All other chemicals, unless stated otherwise, were purchased from Sigma Chemical Co. (St. Louis, MO). The MICs of quinolones were determined by the agar dilution method (21), except that the bacteria were incubated at 43°C, and the agar was minimal medium with thymidine. All experiments were performed at least twice.
Cloning of gyrA genes into pBAD24.
For the cloning of wild-type S. aureus gyrB-gyrA tandem genes, their entire coding region (GenBank accession number D10489) was amplified from strain ISP794 (8325 pig-131) (28) with upstream and downstream primers WTFR2 and WTRV2, respectively (Table 1), which containing engineered NcoI and SphI sites, respectively, to generate pBAD24-29GyrBA. For the cloning of wild-type E. coli gyrA, the gene (GenBank accession number X06744) was amplified from wild-type strain KL16 with the upstream and downstream primers ECOLF1 and ECOLR1, respectively (Table 1), which contained engineered NheI and SphI sites, respectively, to generate pBAD24-1KL16. For the cloning of E. coli gyrA with a Ser83Leu mutation, we used the overlap extension PCR method (26) with KL-16 gyrA cloned in pBAD24 as the template. The left fragment was amplified with primers ECOLF1 and ECOLR2; and the right fragment was amplified with primers ECOLF3 and ECOLR3, the latter of which contains an engineered PstI site (Table 1), to generate pBAD24-S83L. Chimera 7, in which DNA sequences encoding amino acids 84 to 101 of E. coli GyrA (AVYDTIVRMAQPFSLRYM) were replaced with DNA sequences encoding amino acids 85 to 102 of S. aureus GyrA (SIYEAMVRMAQDFSYRYP), was constructed from KL-16 in two steps by using the overlap extension PCR method. We first constructed chimera 6; the left fragment was amplified with ECOLF1 and CHIM6R1, and the right fragment was amplified with CHIM6F1 and ECOLR1. Two fragments were then amplified from chimera 6 (the left fragment was constructed with primers ECOLF1 and CHIM7R, and the right fragment was constructed with primers CHIM7F and ECOLR3) to generate pBAD24-chimera 7. Amplified genes cloned in pBAD24 were electroporated into E. coli DH5α (GIBCO-BRL). Electrotransformants were selected on ampicillin-containing agar, and the insert DNA was then sequenced to confirm its integrity. DNA sequencing was performed by using Taq DyeDeoxy Terminator (Applied Biosystems) with the ABI 3700 PRISM automated sequencer (Massachusetts General Hospital Core Facility). The constructs were then transformed into E. coli KNK453, which was then maintained at 30°C.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| WTFR2 | 5′-CATATTCCATGGTGACTGCATTGTCAGATG |
| WTRV2 | 5′-CATAATGCATGCTTATTATTCTTCATCTGATGATTGTTG |
| ECOLF1 | 5′-AAATTATAGCTAGCAGGAGGGCGGTTAGATGAGCGACCTTGCGAG |
| ECOLR1 | 5′-CATAATGCATGCTTATTCTTCTTCTGGCTCGTCGTCAACGTC |
| ECOLR2 | 5′-GATCGTGTCATAGACCGCCAAGTCACCATGGGGATGGTAT |
| ECOLF3 | 5′-ATACCATCCCCATGGTGACTTGGCGGTCTATGACACGATC |
| ECOLR3 | 5′-GCAGTTTTCGCTTCTGCAGGCGTCGGCGC |
| CHIM6F1 | 5′-TCGTCTATTTATGAAGCAATGGTCCGCATGGCGCAGCCATTC |
| CHIM6R1 | 5′-GACCATTGCTTCATAAATAGACGAGTCACCATGGGGATGGTATTTACCG |
| CHIM7F | 5′-GATTTCTCGTATCGTTATCCGCTGGTAGACGGTCAGGGTAACTTC |
| CHIM7R | 5′-CGGATAACGATACGAGAAATCCTGCGCCATGCGGACCATTG |
| ECOLF9 | 5′-CATGGATCCAGCGACCTTGCGAGAGAAATTACAC |
| ECOLR4 | 5′-CATGGAATTCTTATTCTTCTTCTGGCTCGTCGTCAA |
Cloning of S. aureus gyrA and gyrB, E. coli gyrA and gyrB, and chimera 7 gyrA genes for protein expression.
Cloning of S. aureus ISP794 gyrB into pTrcHisB was performed as described previously (14). S. aureus gyrA was cloned on the backbone of our previously reported vector, pSAGA1 (29), to generate pSAGA3 (29a). Cloning of the complete genes of E. coli gyrA and gyrB separately into pTrcHisA was performed as described previously (31). Cloning of the gyrase-coding region of chimera 7 in pTrcHisA was carried out in a manner similar to that described for E. coli gyrA, except that the upstream and downstream primers used were primers ECOLF9 and ECOLR4, respectively, which contained engineered BamHI and EcoRI sites, respectively.
Overexpression and purification of gyrase subunits.
Overexpression and purification of S. aureus GyrB and E. coli GyrA and GyrB were performed as described previously (14, 31). Chimera 7 GyrA was expressed and purified with an N-terminal six-histidine tag, as in wild-type E. coli GyrA. pSAGA3 (S. aureus gyrA cloned into a modified pBAD/Thio-TOPO vector) was transformed into TOP10 E. coli (Invitrogen, Carlsbad, CA). The fusion protein expressed was then cleaved with recombinant Tobacco Etch virus (rTEV) protease (Invitrogen), and after dialysis, it was reapplied on a nickel-chelate affinity column to adsorb any remaining noncleaved fusion protein and the rTEV protease to yield a pure GyrA subunit (29a).
Gyrase catalytic and DNA cleavage assays.
One unit of gyrase was defined as the amount required to produce half-maximal supercoiling of 0.5 μg of relaxed pBR322 plasmid DNA under the specified test conditions. Two units of each of the subunits, GyrA plus GyrB, was preincubated together for 30 min on ice to reconstitute gyrase holoenzyme. DNA supercoiling activity was assayed with relaxed pBR322 DNA (0.5 μg; Topogen, Columbus, OH) as the substrate. The reaction mixture (20 μl) for E. coli gyrase contained 17.5 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 1.8 mM spermidine, 19 mM KCl, 5 mM dithiothreitol, 1.4 mM ATP, 360 μg bovine serum albumin per ml, 8 μg tRNA per ml, 5% (vol/vol) glycerol, and various concentrations of K+-glutamate and ciprofloxacin. The reaction mixture for S. aureus gyrase was published previously (29). The reaction was carried out at 30°C for 30 min.
DNA cleavage assays were carried out as described above for the catalytic assays, except that ATP was omitted and the DNA substrate used was negatively supercoiled pBR322 DNA (New England Biolabs, Beverly, MA) at a concentration of 16 μg/ml. After 30 min, 2 μl of both sodium dodecyl sulfate (5%) and proteinase K (1 mg/ml) was added, and the incubation was continued for an additional 30 min at 37°C. All experiments were performed at least three times.
The reactions were stopped by adding a mixture of EDTA (50 mM), bromphenol blue, and glycerol. All 20 μl of each reaction mixture was loaded onto a 1.2% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) and run at 3.5 V/cm for 14 to 16 h. The gels were stained with 0.6 μg ethidium bromide per ml for 60 min, destained in water, and scanned and visualized under UV light with FOTO/Analyst (Fotodyne Inc., Hartland, WI). The relative densities of the DNA bands were determined and normalized with Phoretix 1D Quantifier software (Nonlinear; Phoretix, Newcastle upon Tyne, United Kingdom).
RESULTS
Purification and characterization of gyrase proteins.
All proteins were obtained in soluble form at >95% homogeneity (data not shown). The specific activities for DNA supercoiling for gyrase subunits in the presence of an excess of the cognate subunit are shown in Table 2. As shown previously, S. aureus gyrase required supplemental K+-glutamate for supercoiling activity (3, 10); no supercoiling could be demonstrated without K+-glutamate, and the S. aureus gyrase specific activity increased fourfold when the K+-glutamate concentration was increased from 250 mM to 750 mM. Wild-type E. coli GyrA and chimera 7 GyrA in the presence of excess E. coli GyrB manifested similar specific activities of 1.0 × 106 U/mg and 8 × 105 U/mg, respectively, as measured in E. coli buffer. In contrast to S. aureus gyrase, with the addition of K+-glutamate, the specific activities of the enzymes reconstituted with E. coli wild-type GyrA and chimera 7 GyrA decreased progressively and to the same extent, twofold and fourfold, with 100 mM and 700 mM K+-glutamate, respectively (Table 2). Of note, although the intensities of the supercoiled DNA bands were greater when K+-glutamate was added, the specific activity, as determined by titration to half-maximal supercoiling, decreased. We further assessed the two enzymes for potential differences in their kinetics. For these experiments, incubations were performed at room temperature, and the rates of accumulation of the most highly supercoiled DNA band on gels with 0 mM and 300 mM K+-glutamate were similar for the two enzymes over 60 min.
TABLE 2.
Specific activities of recombinant gyrase enzymes
| Assayed subunit | Sp act (Ua/mg) at K+-glutamate concn of:
|
|||||
|---|---|---|---|---|---|---|
| 0 mM | 100 mM | 250 mM | 300 mM | 700 mM | 750 mM | |
| S. aureus GyrAb | 0 | 4.3 × 105 | 1.2 × 106 | |||
| E. coli GyrAc | 1 × 106 | 5 × 105d | NDe | 2.5 × 105d | ||
| Chimera 7 GyrAc | 7.9 × 105 | 4 × 105 | 3 × 105 | 2 × 105 | ||
U, unit; 1 unit of gyrase was defined as the amount required to produce half-maximal supercoiling of 0.5 μg of relaxed pBR322 plasmid DNA under the specified test conditions.
Reconstituted with wild-type S. aureus GyrB.
Reconstituted with wild-type E. coli GyrB.
The specific activity of KL16 GyrA was not limited by the activity of the GyrB subunit at the different K+-glutamate concentrations tested.
ND, not determined.
Activity of ciprofloxacin against purified wild-type E. coli gyrase and chimera 7 gyrase.
To determine quinolone inhibition of wild-type and chimeric gyrases, we measured the extent of inhibition of gyrase-mediated negative supercoiling of relaxed pBR322 DNA. Without K+-glutamate, the 50% inhibitory concentrations (IC50s) of the chimera 7 gyrase (chimera 7 GyrA reconstituted with wild-type GyrB) were 0.62 to 1.25 μg/ml, which were fourfold higher than those of wild-type gyrase, 0.16 to 0.31 μg/ml, with the latter value being similar to that published previously (1) (Table 3). Upon addition of 100 mM K+-glutamate, the IC50 of wild-type gyrase increased fourfold, whereas the IC50 of chimera 7 gyrase decreased fourfold. With a K+-glutamate concentration of 300 mM, the IC50 of wild-type gyrase did not change further, whereas the IC50 of the chimera 7 gyrase increased fourfold. In order to be certain that the differences in enzyme sensitivities to ciprofloxacin were not due to differences in the activities of the gyrase subunits, the experiments were repeated in the presence of fourfold excesses of the GyrA subunits, and the same results were found. Thus, at maximal activities, wild-type and chimera 7 GyrA conferred different sensitivities to ciprofloxacin.
TABLE 3.
Ciprofloxacin inhibition of gyrase DNA supercoiling activity and stimulation of cleavage complex formation with gyrase
| Recombinant enzyme | IC50 (μg/ml) with the following K+-glutamate concn:
|
CC50 (μg/ml) with the following K+-glutamate concn:
|
||||
|---|---|---|---|---|---|---|
| 0 mM | 100 mM | 300 mM | 0 mM | 100 mM | 300 mM | |
| E. coli GyrA2 GyrB2 | 0.16-0.31 | 0.62-1.25 | 0.62-1.25 | 0.31 | 0.16-0.31 | 0.62 |
| Chimera 7 GyrA2 and E. coli GyrB2 | 0.62-1.25 | 0.16-0.31 | 0.62-1.25 | 0.62-1.25 | 1.25 | 5 |
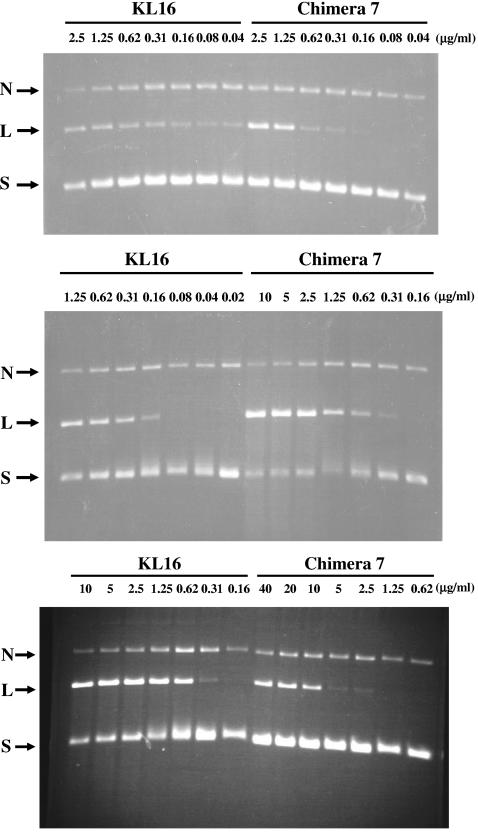
To compare further the relative sensitivities of the wild-type and the chimera 7 gyrases to ciprofloxacin and the effect of K+-glutamate on drug sensitivity, we used the cleavage complex (CC) formation assay. In the absence of K+-glutamate, the potency of ciprofloxacin in stimulating half-maximal intensity of linear DNA cleavage complex formation (CC50) was similar to or twofold less with chimera 7 gyrase relative to that with wild-type gyrase (Table 3) (Fig. 1, top panel). At K+-glutamate concentrations of 100 mM and 300 mM, four- and eightfold greater concentrations of ciprofloxacin, respectively, were needed to stimulate half-maximal formation of cleavage complexes with chimera 7 gyrase than with wild-type gyrase (Fig. 1, middle and bottom panels). The CC50 of wild-type gyrase was the lowest with a K+-glutamate concentration of 100 mM and changed little with K+-glutamate concentrations of 0 mM to 300 mM. With chimera 7 gyrase, however, there was a progressive increase in the ciprofloxacin CC50 with increasing concentrations of K+-glutamate. Thus, chimera 7 gyrase was more resistant to ciprofloxacin than wild-type gyrase for both inhibition of catalytic activity and stimulation of cleavage complexes. This difference in resistance, however, was more evident in the presence of K+-glutamate.
FIG. 1.
Effect of K+-glutamate on ciprofloxacin-mediated DNA cleavage by E. coli wild-type and chimera 7 gyrases. The ciprofloxacin concentration is indicated. N, L, and S, nicked, linear, and supercoiled DNA, respectively. The K+-glutamate concentration in the reactions were (top) 0 mM, (middle) 100 mM, and (bottom) 300 mM.
gyrA complementation assays.
Because the activities of the wild-type and the chimeric enzymes differed and were dependent on the K+-glutamate concentration, we next determined the activity of ciprofloxacin against cells of an E. coli thermosensitive gyrA mutant expressing the plasmid-encoded GyrA subunits at a nonpermissive temperature.
E. coli gyrA+, E. coli gyrA (S83L), S. aureus gyrB-gyrA tandem genes, and chimera 7 gyrA genes were cloned into pBAD24 and transformed into KNK453 gyrA(Ts). The pBAD24 vector was also transformed into KNK453 to serve as a control for complementation on the same ampicillin-containing plate. We cloned S. aureus gyrB in tandem with gyrA because S. aureus GyrA cannot form an active holoenzyme with E. coli GyrB (J. Strahilevitz and D. C. Hooper, unpublished data), and the two genes in S. aureus are transcribed as a polycistronic RNA (13). We expressed pBAD24, which encodes the two genes, in DH5α cells and validated that the two proteins were translated separately by the sizes of the bands on a sodium dodecyl sulfate-polyacrylamide gel. Thus, this construct allowed the potential formation of active S. aureus gyrase or a heterologous S. aureus GyrA-E. coli GyrB gyrase in KNK453 cells. All transformants grew at the permissive temperature (30°C) on ampicillin-containing agar. At the restrictive temperature (42°C), cells containing pBAD24 with wild-type gyrA, pBAD24 with gyrA with the S83L mutation, and pBAD24 with chimera 7 gyrA were able to grow to the same degree. Cells containing pBAD24 and pBAD24 with 29GyrBA, however, failed to grow at the restrictive temperature. Thus, S. aureus GyrA appears to be unable to complement a thermosensitive E. coli GyrA, but an E. coli GyrA with a replaced extended α4 domain of S. aureus functioned similarly to wild-type GyrA in vivo.
Activities of quinolones against KNK453 transformants in the presence and the absence of supplemental K+-glutamate.
We first determined the MIC of ciprofloxacin against the E. coli gyrA and chimera 7 gyrA transformants of KNK453 on minimal medium. The MICs of the three transformants at the permissive temperature 30°C were the same. At the restrictive temperature (43°C), the MIC of ciprofloxacin was 0.02 μg/ml and was the same for cells containing pBAD24 with wild-type gyrA and pBAD24 with chimera 7 gyrA, but the MIC of ciprofloxacin for pBAD24 with gyrA with the S83L mutation was increased fourfold (Table 4). Thus, although mutant E. coli GyrA (S83L) expresses resistance in E. coli cells under these conditions, chimera 7 GyrA did not confer the increased resistance to quinolones that was demonstrated with the purified chimeric enzyme.
TABLE 4.
Activities of ciprofloxacin against KNK453 (gyrAts) transformed with pBAD24-gyrase genes on different agar mediaa
| Plasmid | Ciprofloxacin MIC (μg/ml) in the following growth mediab:
|
|||
|---|---|---|---|---|
| I | II | III | IV | |
| KL16 gyrAc | 0.02 | 0.04 | 0.04 | 0.06 |
| S83Ld | 0.08 | 0.08-0.16 | 0.08-0.16 | 0.08-0.16 |
| Chimera 7e | 0.02 | 0.08 | 0.04 | 0.06 |
Determined by agar dilution method at 43°C.
I, minimal medium; II, medium I plus K+-glutamate 300 mM; III, minimal medium with Casamino Acid supplementation; IV, medium III plus K+-glutamate at 300 mM.
Wild-type E. coli gyrA.
KL16 Ser83Leu gyrA.
Chimera of E. coli and S. aureus GyrA.
We next measured the MICs of ciprofloxacin against KNK453 with various plasmid constructs at 43°C on minimal medium supplemented with amino acids. Supplementation with K+-glutamate as the sole source of amino acids resulted in a slight increase in the MICs of ciprofloxacin, to values of 0.04 μg/ml for cells with pBAD24 with wild-type gyrA and 0.08 to 0.16 μg/ml for cells with pBAD24 with gyrA with the S83L mutation. The MIC of ciprofloxacin for cells with pBAD24 with the chimera 7 gyrA, however, increased fourfold to 0.08 μg/ml and was consistently twofold higher than that for cells containing pBAD24 with wild-type gyrA. The same results were obtained with K+-glutamate at 150 mM, as well as in minimal medium supplemented with K+-aspartate (300 mM). No difference in ciprofloxacin MICs for pBAD24 with the wild-type gyrA and pBAD24 with the chimera 7 gyrA was observed when K+-glutamate was replaced with glycine or leucine. Similarly, supplementation with Casamino Acids resulted in MICs of ciprofloxacin of 0.04 μg/ml for both cells with pBAD24 with the wild-type gyrA and cells with pBAD24 with chimera 7 gyrA and 0.08 to 0.16 μg/ml for cells with pBAD24 with gyrA with the S83L mutation. Thus, supplementation of the medium with acidic amino acids alone but not neutral or mixed amino acids allowed the in vivo expression of low-level ciprofloxacin resistance conferred by chimera 7 GyrA.
DISCUSSION
The differential sensitivity of E. coli and S. aureus gyrases to quinolones is thought to be responsible for the difference in the primary enzyme target in the two species (3), a property that can potentially affect the outcomes of drug action (6, 15). Mutations in the α4-helix domain of both GyrA subunits are pivotal in determining quinolone resistance, and this domain differs between the wild-type enzymes of the two species. Therefore, we hypothesized that the α4-helix domain of S. aureus is responsible for the native increased resistance of S. aureus gyrase to quinolones. In support of this hypothesis, we have demonstrated that an E. coli GyrA subunit containing the α4-helix domain of S. aureus confers a higher IC50 of ciprofloxacin to gyrase reconstituted with wild-type GyrB than wild-type GyrA does, and the chimeric enzyme exhibited specific activity for DNA supercoiling similar to that of the wild-type enzyme in vitro and was functional in vivo in complementing a thermosensitive E. coli GyrA. Unexpectedly, the chimeric enzyme in E. coli cells expressed no resistance under conditions in which expression of a GyrA S83L subunit conferred the expected resistance known to be mediated by this mutant subunit (18), suggesting that the conditions within the E. coli cytoplasm may have obviated the properties of drug resistance of the chimeric GyrA found in vitro.
The macromolecular composition of S. aureus is broadly similar to that of E. coli (33). In contrast to E. coli, however, in S. aureus the acidic amino acids glutamate and aspartate are the amino acids present at the highest concentrations in the cytoplasmic pool (33), and when the two species are grown in a similar minimal medium, the concentration of glutamate is at least eightfold higher in S. aureus than in E. coli (2, 19). Thus, S. aureus topoisomerases may have evolved to accommodate higher glutamate concentrations. The concentration of K+-glutamate (and the concentrations of other potassium salts of the dicarboxylic α-amino acids glutamate and 2-methylglutamate) affects the specific activity of S. aureus gyrase and topoisomerase IV (3). Addition of K+-glutamate at a concentration of 700 mM (in excess of its intracellular concentration) increased the specific activity of S. aureus gyrase from 1 × 103 to 5 × 105 U/mg, a value similar to that for E. coli gyrase in the absence of glutamate (3). We found that the specific activity of S. aureus GyrA increased from undetectable to 4.3 × 105 U/mg with the introduction of K+-glutamate at 250 mM and increased threefold more at a K+-glutamate concentration of 750 mM. The quinolone resistance of purified S. aureus gyrase has thus, of necessity, been measured in the presence of glutamate. Thus, we postulated that the resistance conferred by the extended α4 domain of chimera 7 GyrA might be affected by glutamate.
DNA supercoiling by gyrase constituted with chimera 7 GyrA behaved like gyrase constituted with wild-type GyrA in response to supplemental glutamate, and the gyrases had similar specific activities that declined similarly with increasing glutamate concentrations. In contrast, enzyme sensitivity to ciprofloxacin was differentially affected by glutamate. Although the effect of the glutamate concentration on IC50 was complex, the overall pattern for both the IC50 and the CC50 data indicated increasing ciprofloxacin resistance with increasing concentrations of K+-glutamate. The DNA cleavage complex assay, which is thought to reflect most closely the relevant cytotoxic actions of quinolones, demonstrated similar potencies for ciprofloxacin for chimera 7 gyrase and wild-type gyrase in the absence of glutamate. With the addition of K+-glutamate, however, the differential behavior of the wild-type and the chimera 7 gyrases was evident, with eightfold more ciprofloxacin required to stimulate half-maximal cleavage complexes with chimera 7 gyrase than with wild-type gyrase over a range of glutamate concentrations. Thus, in reaction mixtures with glutamate concentrations that approximate those in the cytoplasm of S. aureus, we observed a further increase in resistance of the chimera 7 gyrase to ciprofloxacin. Nevertheless, the CC50 of ciprofloxacin with wild-type S. aureus gyrase, determined with a similar K+-glutamate concentration of 250 mM, was 6- to 10-fold (30 to 50 μg/ml) higher than that with the chimera 7 gyrase, indicating that the decreased sensitivity of S. aureus to quinolones cannot be solely explained by the differences in the extended α4-helix domain of GyrA between S. aureus and E. coli.
The effect of glutamate on S. aureus gyrase can vary among assays (3, 10). Hiasa et al. have compared the catalytic and noncatalytic activities of S. aureus and E. coli gyrases (10) and showed that the requirement for a high glutamate concentration (400 mM) was unique for S. aureus gyrase-mediated wrapping of DNA and was a prerequisite for quinolone-mediated replication fork arrest in vitro. Thus, if a high concentration of glutamate was not reached in vivo, ternary complexes of gyrase-DNA-quinolone would not form to arrest the replication fork, which explains why gyrase is a less favorable target in S. aureus (10). These differences between E. coli and S. aureus gyrases, however, do not explain why topoisomerase IV becomes the primary target in S. aureus. It is possible that modifications of S. aureus gyrase that enable its functionality (i.e., wrapping of DNA) in the presence of an increased concentration of glutamate lead to associated changes in the extended α4-helix domain that are associated with increased resistance to quinolones. It is also possible that the increase in ciprofloxacin resistance manifested in the cleavage complex assays with increasing concentrations of glutamate reflects a decrease in the stability of DNA binding and complex formation for the chimeric enzyme.
Because glutamate supplementation increased the difference in ciprofloxacin resistance between purified chimera 7 gyrase and wild-type gyrase, we attempted to augment the resistance of chimera 7 gyrase in vivo by supplementation of the growth medium with glutamate. Remarkably, supplementation with high concentrations of glutamate (300 mM) or aspartate singly but not neutral amino acids or Casamino Acids (which contain a lower concentration of glutamate [14 mM] [9]) unmasked a difference in resistance to ciprofloxacin between complementing chimera 7 gyrase and wild-type gyrase expressed in E. coli cells. The concentration of glutamate in S. aureus cells during exponential growth is approximately 77 to 88 mM (2, 17), which is a concentration lower than that used to optimize the enzyme assays for S. aureus but one that is 10-fold higher than that in E. coli, 7.8 mM (19). Thus, a relatively higher glutamate concentration is likely a biologically relevant difference between the cytoplasmic milieus in which gyrase functions in E. coli and S. aureus. It is known that further supplementation of the amino acids in rich media such as nutrient broth does not affect the intracellular amino acid concentration in orders of magnitude (2, 19). Although it is uncertain whether high-level supplementation of the medium has direct effects on the cytoplasmic concentrations of these amino acids, the findings establish that the quinolone resistance conferred by the extended α4 domain of S. aureus can be expressed in E. coli cells in vivo under certain growth conditions.
The extended α4-helix domain that was exchanged in chimera 7 GyrA encompasses the quinolone resistance-determining region, in which the majority of the resistance mutations in gyrA selected with quinolones generally cluster (4, 35). E. coli gyrase and S. aureus gyrase share 48% amino acid identity, and in the α4-helix domain 5 of 12 amino acids differ. In addition, of the 50 amino acids in the extended region downstream of the α4 helix that was studied, 2 amino acids differ: a proline positioned 5 and 12 residues after the α4 helix in E. coli and S. aureus, respectively and a leucine in E. coli that aligns with a tyrosine in S. aureus positioned nine amino acids after the α4 helix. This extended region is predicted to be distinct from the α4 helix, as modeled in the crystal structure of an E. coli GyrA fragment (20), and has not been the site of mutations conferring quinolone resistance; but because of the differences between species, we elected to include this region in chimera 7. Because the crystal structure of S. aureus gyrase has not yet been solved, it is unclear what part of the replaced region participates in the active-site pocket of the enzyme and what segment is involved in quinolone binding. Our data thus support prior mutational data that have indicated that the α4 helix is important in the determination of the quinolone sensitivity of gyrase and may contribute to the differences in the intrinsic reduced sensitivities of S. aureus gyrase. In addition, this domain also contributes to the modulation of quinolone sensitivity in the presence of glutamate, thereby reflecting the additive effects of the target enzyme structure and the cellular milieu in fostering the intrinsic quinolone resistance of S. aureus gyrase. Further studies will be necessary to determine which of the amino acid differences in the extended α4-helix domain are responsible for its ability to confer increased resistance to quinolones and responsiveness to glutamate.
In summary, we have established the contribution of the α4-helix domain of gyrase to the difference in quinolone susceptibility between E. coli and S. aureus. The results show that replacement of the extended α4-helix domain of E. coli GyrA with that of S. aureus GyrA results in an active enzyme that confers quinolone resistance in vitro and in intact E. coli cells. The results also show that the level of this resistance is modulated by glutamate, an amino acid that differs in its cytoplasmic concentration between E. coli and S. aureus cells.
Acknowledgments
We thank Marcia Goldberg for providing pBAD24 and Mordehay Vatury for helpful discussions.
This work was supported in part by a fellowship from American Physicians Fellowship for Medicine in Israel (to J.S.) and by grants from the U.S. Public Health Service, National Institutes of Health (grant AI 23988 to D.C.H.).
REFERENCES
- 1.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björklind, A., and S. Arvidson. 1978. Influence of amino acids on the synthesis of an extracellular proteinase from Staphylococcus aureus. J. Gen. Microbiol. 107:367-375. [Google Scholar]
- 3.Blanche, F., B. Cameron, F. X. Bernard, L. Maton, B. Manse, L. Ferrero, N. Ratet, C. Lecoq, A. Goniot, D. Bisch, and J. Crouzet. 1996. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob. Agents Chemother. 40:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helena da Cruz, S., E. M. Cilli, and J. R. Ernandes. 2002. Structural complexity of the nitrogen source and influence on yeast growth and fermentation. J. Inst. Brewing 108:54-61. [Google Scholar]
- 10.Hiasa, H., M. E. Shea, C. M. Richardson, and M. N. Gwynn. 2003. Staphylococcus aureus gyrase-quinolone-DNA ternary complexes fail to arrest replication fork progression in vitro—effects of salt on the DNA binding mode and the catalytic activity of S. aureus gyrase. J. Biol. Chem. 278:8861-8868. [DOI] [PubMed] [Google Scholar]
- 11.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31:S24-S28. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, D. C. 2003. Mechanisms of quinolone resistance, p. 41-67. In D. C. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents. ASM Press, Washington, D.C.
- 13.Ince, D., and D. C. Hooper. 2003. Quinolone resistance due to reduced target enzyme expression. J. Bacteriol. 185:6883-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of garenoxacin (BMS-284756, T3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 16.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koujima, I., H. Hayashi, K. Tomochika, A. Okabe, and Y. Kanemasa. 1978. Adaptational change in proline and water content of Staphylococcus aureus after alteration of environmental salt concentration. Appl. Environ. Microbiol. 35:467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx, D., B. Flouret, and J. Van Heijenoort. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, p. 1. In Approved standard M7-A6, vol. 23. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 22.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship of the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, X. S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 25.Pfeiffer, E. S., and H. Hiasa. 2004. Replacement of ParC alpha4 helix with that of GyrA increases the stability and cytotoxicity of topoisomerase IV-quinolone-DNA ternary complexes. Antimicrob. Agents Chemother. 48:608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19-1-e19-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, H. J., K. A. Nichol, D. J. Hoban, and G. G. Zhanel. 2002. Dual activity of fluoroquinolones against Streptococcus pneumoniae: the facts behind the claims. J. Antimicrob. Chemother. 49:893-895. [DOI] [PubMed] [Google Scholar]
- 28.Stahl, M. L., and P. A. Pattee. 1983. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J. Bacteriol. 154:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strahilevitz, J., and D. C. Hooper. 2005. Dual targeting of topoisomerase IV and gyrase to reduce mutant selection: direct testing of the paradigm by using WCK-1734, a new fluoroquinolone, and ciprofloxacin. Antimicrob. Agents Chemother. 49:1949-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Strahilevitz, J., Y. Onodera, and D. C. Hooper. 2005. An improved expression plasmid for affinity purification of Staphylococcus aureus gyrase A subunit. Protein Expr. Purif. [Epub ahead of print.] [DOI] [PubMed]
- 30.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trucksis, M., J. S. Wolfson, and D. C. Hooper. 1991. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 173:5854-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 34.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, H., M. Nakamura, M. Bogaki, H. Ito, T. Kojima, H. Hattori, and S. Nakamura. 1993. Mechanism of action of quinolones against Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 37:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, X. L., C. Xu, J. Domagala, and K. Drlica. 1997. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc. Natl. Acad. Sci. USA 94:13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]