Abstract
Two erg3 genes encoding C-5 sterol desaturase enzymes (Erg3A and Erg3B) in Aspergillus fumigatus were characterized with respect to their nucleotide sequences and null mutant phenotypes. Targeted disruption of the erg3A and erg3B genes and a double gene knockout, erg3A− erg3B−, showed that they are not essential for A. fumigatus viability. Mutant phenotypes clearly showed that Erg3B is a C-5 sterol desaturase, but no apparent role for Erg3A in A. fumigatus ergosterol biosynthesis was found. Susceptibility to amphotericin B, itraconazole, fluconazole, voriconazole, and ketoconazole was not altered in isolates in which erg3A and erg3B were knocked out alone and in combination.
Aspergillus fumigatus has become the most prevalent airborne fungal pathogen in developed countries, causing allergic diseases, fungal balls, and fatal invasive aspergillosis (27). Furthermore, the incidence of aspergillosis has increased in recent years, due primarily to an increase in immunocompromised patients, particularly those with acute leukemia and bone marrow or solid organ transplants (25).
The majority of A. fumigatus isolates are susceptible in vitro to itraconazole, voriconazole, and amphotericin B (AmB) (16, 25); however, isolates with itraconazole MICs of >8 μg/ml have already been well documented and categorized as resistant (4, 9, 12, 25, 33). In addition, those isolates with high MICs have been correlated with poor clinical outcomes in both animal models and patients (10, 11). In contrast, high MICs to AmB are uncommon, although therapeutic failures with this polyene are frequently reported (22).
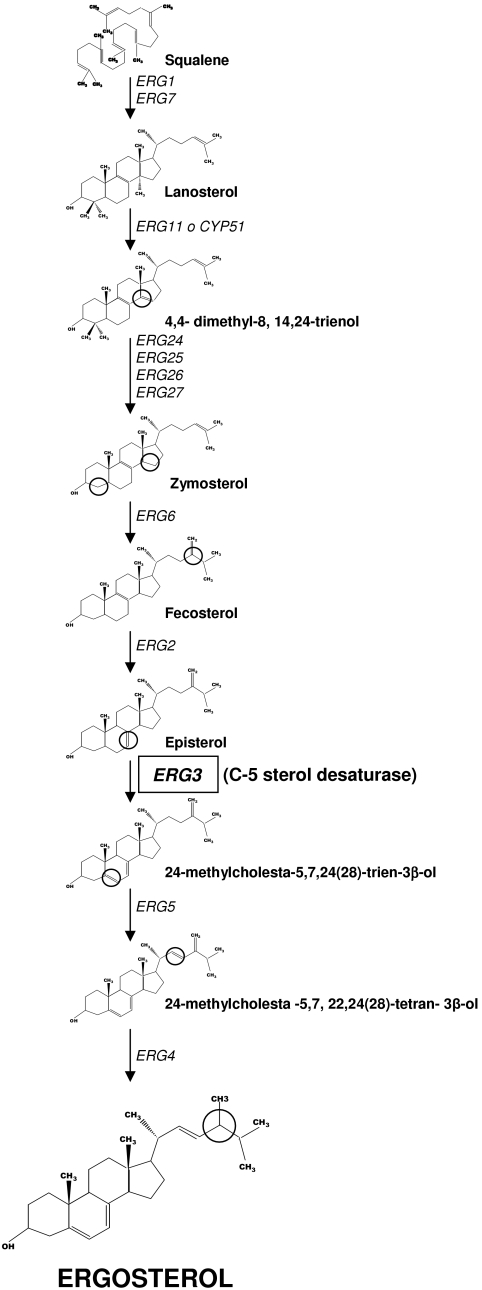
Azole drugs inhibit the 14-α sterol demethylase (Erg11/Cyp51) in the ergosterol biosynthesis pathway (Fig. 1). In A. fumigatus, azole drug resistance has been matched with different types of resistance mechanisms: (i) alteration of the cellular target, such as mutations that decreases the affinity of the enzyme for azole compounds, and (ii) resistance due to altered drug transport (12, 28, 31, 35, 44). In addition, other resistance mechanisms related to modification of other enzymes involved in ergosterol biosynthesis have been described for yeasts. One such enzyme is the C-5 sterol desaturase encoded by erg3. Mutations or inactivation of the erg3 gene in Saccharomyces cerevisiae have been associated with azole and polyene drug resistance (3, 20, 44). The sterol composition of Candida albicans clinical isolates exhibiting azole and amphotericin B resistance has shown an accumulation of ergosta-7-22-dienol, ergosta-7-enol, and episterol, which are features of the absence of C-5 sterol desaturase activity (23, 24). Furthermore, the loss of function of the erg3 gene in Candida dubliniensis has been described as the primary mechanism of generated itraconazole resistance in vitro (37). Also, a reduction of the expression of the erg3 gene in Candida lusitaniae has been found in two amphotericin B-resistant isolates (50). However, other authors have reported that C. albicans isolates with erg3 deleted and Candida glabrata erg3 null mutants remain susceptible in vitro to amphotericin B (15, 43). Despite all this previous work on yeasts, no study has been done on A. fumigatus. This work describes the identification of three genes in A. fumigatus (erg3A, erg3B, and erg3C) encoding putative C-5 sterol desaturases and the construction of single (erg3A and erg3B) and double (erg3A− erg3B−) mutant strains lacking functional copies of the genes and their phenotypic characterization.
FIG. 1.
MATERIALS AND METHODS
Strains.
The CM-237 strain of A. fumigatus (mold collection of the Spanish National Center for Microbiology) was used throughout this work (30). The fungi were grown at 37°C in GYEP (2% glucose, 0.3% yeast extract, 1% peptone), potato dextrose agar (Oxoid, Madrid, Spain), RPMI (Angus; Oxoid), or minimal medium (6). For propagation of plasmids, Escherichia coli strain JM109 was grown in Luria-Bertani (LB) medium (41) supplemented with ampicillin (100 μg/ml).
Cloning and DNA sequencing.
The erg3 fragments were PCR amplified and cloned into the pGEM-T Easy vector system (Promega, Madrid, Spain). DNA inserts of recombinant plasmids were sequenced with the BigDye terminator cycle sequencing ready reaction system (Applied Biosystems, Madrid, Spain) according to the manufacturer's instructions. Sequence analysis was performed on an ABI Prism 377 DNA sequencer (Applied Biosystems), using the sequencing facilities available in the Genomics Department at the Instituto de Salud Carlos III, Majadahonda, Madrid, Spain.
Primer design and PCR conditions.
The initial sequence used was a 775-bp sequenced tag (af51C7) from an A. fumigatus clone library (ID3355) that was obtained as a gift from J. P. Latge (Pasteur Institute, Paris, France). This sequence showed high homology with different C-5 sterol desaturases encoded by erg3.
Primers designed to cover the full genomic sequence from each gene were used to PCR amplify each of the corresponding sequences from the A. fumigatus CM-237 genomic DNA (Table 1). All the primers used in this study were synthesized by Sigma Genosys (Madrid, Spain). PCRs were carried out in a 50- to 100-μl volume containing 1× PCR buffer (Applied Biosystems); 2 mM MgCl2 (Applied Biosystems); 250 μM each of dATP, dGTP, dCTP, and dTTP (Applied Biosystems); 1 μM of each primer; 2.5 units of Taq DNA polymerase (Applied Biosystems); and 25 to 50 ng of A. fumigatus genomic DNA. Amplification was performed in a thermal cycler (GeneAmp PCR system 9700; Applied Biosystems) for one cycle of 5 min at 94°C and then 30 cycles of 30 s at 94°C, 45 s at the necessary melting temperature, and 1 to 2 min at 72°C, followed by one final cycle of 10 min at 72°C. The PCR products were analyzed by electrophoresis on agarose gels.
TABLE 1.
Oligonucleotide primers used in this work
| Primer | Orientation, 5′→3′ | Sequence | Use |
|---|---|---|---|
| ERG3C | Antisense | GCAGGCGTTTGTAGATGG | Specific for erg3A |
| ERG3D | Sense | ATGGACGTTGTGCTTGAC | Specific for erg3A |
| ERG3F | Sense | ATACCTACCTTCAGTACC | Specific for erg3A |
| ERG3Z | Antisense | CTCAGGAAGTTTTCTTGG | Specific for erg3A |
| ERG321 | Sense | GCCAAACAATCAAGGCACAG | Specific for erg3A |
| ERG322 | Antisense | GATCTATGGTCGGTTGGTTG | Specific for erg3A |
| ERG3B1 | Sense | CGGCACCATGGATATTG | Specific for erg3B |
| ERG3B2 | Antisense | AGATGTAGGAGAGGGTTG | Specific for erg3B |
| ERG3B3 | Sense | ATACCGTTGCCATTCTATTG | Specific for erg3B |
| ERG3B4 | Antisense | CGCAACGGAATACGATATTG | Specific for erg3B |
| ERG3B5 | Sense | CGAGGAAGTCGATCAAAG | Specific for erg3B |
| ERG3B9 | Sense | ATCCCCATGGAGATTGCG | Specific for erg3B |
| ERG3B10 | Antisense | GTCGTGAATCATTACCGTC | Specific for erg3B |
| ERG3B6 | Sense | GACAGAGTAGGCACGTAG | Specific for erg3B |
| ERG3B7 | Antisense | CCCCTTATCTGGCTGACA | Specific for erg3B |
| ERG3C1 | Sense | ATCCTCACGCTGCCATGG | Specific for erg3C |
| ERG3C2 | Antisense | GTCGCCGTCGTGAATGAG | Specific for erg3C |
| ERG3C3 | Sense | ATGGATGTCGCTCTCGAG | Specific for erg3C |
| ERG3C4 | Antisense | TTGAAGTCCCTCACTCCC | Specific for erg3C |
| ERG3C5 | Sense | AGGATATAAATACCTCCG | Specific for erg3C |
| ERG3C6 | Antisense | CTAATCGTCATGCATATG | Specific for erg3C |
| T-7 | Universal | TAATACGACTCACTATAGGGCGA | Clone sequencing |
| Sp-6 | Reverse | ATTTAGGTGACACTATAGAATAC | Clone sequencing |
| ERG3Akpn1 | Sense | CCTTCAGGTACCCGCTC | Specific for erg3A |
| ERG3Akpn2 | Antisense | GAGCGGGTACCTGAAGG | Specific for erg3A |
| ERG3BKpn1 | Sense | CAAGTTGGTACCGACTCC | Specific for erg3B |
| ERG3BKpn2 | Antisense | GGAGTCGGTACCAACTTG | Specific for erg3B |
| HygKpn1 | Sense | GGTACCTGATATTGAAGG | Specific for hyg |
| HygKpn2 | Antisense | GGTACCTTAACTGGTTCC | Specific for hyg |
| Tub1 | Sense | AACCAAATTGGTGCCGC | Specific for tub1 |
| Tub2 | Antisense | CACGGATCTTGGAGATC | Specific for tub1 |
RNA isolation and RT-PCR.
Total RNA was extracted from the A. fumigatus CM-237 strain, and reverse transcription (RT) reactions were performed (30). Amplification of cDNAs was carried out using the LightCycler PCR system (Roche Diagnostics, Madrid, Spain). The primers ERG3F and ERG3Z were used to amplify the cDNA from erg3A, and primers ERG3B1 and ERG3B2 were used for erg3B (Table 1). The RT-PCR products were resolved by electrophoresis on 1.4% agarose gels and were purified for sequencing. Also, the primer set Tub1-Tub2 (Table 1) was used for amplification of the A. fumigatus β-tubulin housekeeping gene (Tub1) (GenBank accession number AY048754). LightCycler PCRs were set up with FastStart DNA Master SYBR Green (Roche Diagnostic). Each assay was repeated on three separate days. PCR efficiencies were calculated from the curve slopes given by the LightCycler software (Roche Diagnostic).
DNA isolation and hybridization.
Genomic DNAs from the A. fumigatus strains were obtained using a rapid extraction procedure (46) and digested with different restriction enzymes. Southern analysis was performed as previously described (30). Probes for erg3A and erg3B were obtained by restriction digestion of the appropriate clones. The desired fragments were fractionated in 0.7% low-melting-point agarose gels and excised for labeling with a random-prime DNA labeling system (ECL; Amersham Biosciences) according to the manufacturer's instructions.
Computer analyses.
The A. fumigatus genome databank at The Institute for Genomic Research (TIGR) was used to search for A. fumigatus sequences (http://tigrblastn.tigr.org/ufmg/data/blastn-a_fumigatus). The amino acid sequences of putative C-5 sterol desaturase gene fragments were deduced from nucleotide sequences by using the MapDraw software package and analyzed using the MegAlign software package (DNAstar, Inc., Lasergene, Madison, Wis.). The BLASTp program was used to search for homologous sequences of known proteins in the GenBank database. Multiple amino acid alignments were derived by CLUSTAL analysis, and dendrograms were generated by the unweighted pair-group method using arithmetic averages (19). The final phylogeny was produced by applying the neighbor-joining method (40) to the distance and alignment data.
Molecular cloning and construction of disruption vectors.
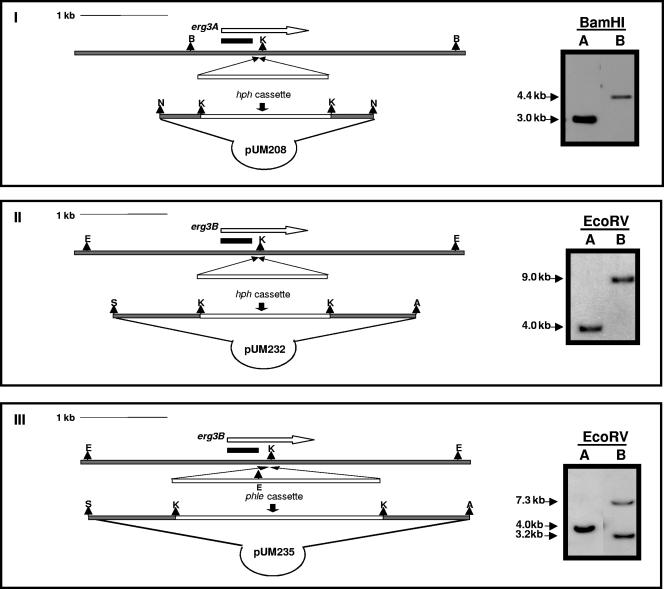
The full coding sequence of erg3A of A. fumigatus was PCR amplified as previously described (with primers ERG3D and ERG3Z) and cloned into a pGEM-T vector system (Promega) to obtain plasmid pUM110. The coding sequence of erg3B, plus 1 kb on each side, was PCR amplified with primers ERG3B6 and ERG3B7 and cloned into a pGEM-T vector system to obtain plasmid pUM226. A KpnI restriction site was engineered in the middle of erg3A and erg3B coding sequences by using PCR site-specific mutagenesis (48) with primers described in Table 1 (ERG3Akpn1 and ERG3Akpn2 for erg3A and ERG3Bkpn1 and ERG3Bkpn2 for erg3B). The hygromycin B (hph) resistance cassette (8) was amplified from plasmid pID621 (kindly provided by D. W. Holden) with primers HygKpn1 and HygKpn2 to add flanking KpnI restriction sites for the construction of the disruption vector. The 1.4-kb hph cassette was inserted in the KpnI restriction site of pUM206 to create pUM208. The 1.4-kb hph cassette was inserted in the KpnI restriction site of pUM226 to create pUM232. The phleomycin (phle) resistance cassette was excised from plasmid pID624 (kindly provided by D. W. Holden) by KpnI digestion and inserted in the KpnI restriction site of pUM226 to create pUM235 (Fig. 2).
FIG. 2.
Diagrams of plasmid constructs used for creating the A. fumigatus erg3A-deficient mutant strain (pUM208) (I), the erg3B− strain (pUM232) (II), and the double erg3A− erg3B− mutant (pUM235) (III). Arrows represent the full coding sequences, gray bars represent the genomic sequence, unfilled boxes indicate the hygromycin resistance cassette (hph) (I and II) or phleomycin resistance cassette (phle) (III), and black bars indicate PCR fragments used as probes. The restriction enzymes used were ApaI (A), BamHI (B), EcoRV (E), KpnI (K), NotI (N), and SacI (S). On the right, Southern hybridization analysis of erg3A− single mutant strain CM-A80 (I), erg3B− strain CM-B866 (II), and the double erg3A− erg3B− strain (III) is shown. Genomic DNAs were digested with BamHI and hybridized using the erg3A gene probe for CM-A80 (I). CM-B866 (II) and CM-AB118 (III) were detected by digesting the genomic DNAs with EcoRV and hybridizing with the erg3B gene probe.
Aspergillus fumigatus transformations.
A. fumigatus transformation experiments were done by electroporation, using a protocol previously described (42) with subsequent modifications (12, 49). Hygromycin B (Sigma, Madrid, Spain) (130 μg/ml) was used for single-transformant selection, and phleomycin (Cayla, Madrid, Spain) (150 μg/ml) was used for double-transformant selection. Mutants were named by a letter(s) from the corresponding gene(s) (A, B, or AB) and number. Genomic DNAs from hygromycin- and phleomycin-resistant transformants and the parental strain were all digested with different restriction enzymes (Amersham Biosciences, Madrid, Spain). Southern analysis was performed to confirm gene target events as previously described (12, 30, 41).
Antifungal susceptibility testing.
Broth microdilution susceptibility testing was performed as described in the CLSI document M38-A (5) with minor modifications (7, 36, 39). Itraconazole and ketoconazole (both from Janssen Pharmaceutical S.A., Madrid, Spain), voriconazole and fluconazole (both from Pfizer S.A., Madrid, Spain), and amphotericin B (Sigma Aldrich Quimica, S.A., Madrid, Spain) were tested. Susceptibility tests were performed at least three times with each strain on different days.
Antifungal susceptibility was also performed by Etest according to the manufacturer's instructions (AB Biodisk, Solna, Sweden).
Sterol analysis.
Total ergosterol of strain CM-237 and its mutants was extracted after 18 h of growth in MM liquid medium at 37°C in an orbital sacker incubator (150 rpm), using the protocol described by Arthington-Skaggs et al. (2). Ergosterol content was analyzed by high-pressure liquid chromatography using a μBondapak C18 column (Waters LC Module I plus; Waters Corporation, Madrid, Spain). The quantities of sterols were calculated with Millenium32 and Millenium32 photodiode array detector software (Waters Corporation). Each experiment was repeated at least three times.
An aliquot of 200 mg of fungal mat was used for sterol extraction to be analyzed by gas chromatography-mass spectrometry (GC-MS). The extraction was performed as described previously (2), but neutral lipids were extracted twice with 1.5 ml hexane. Sterols were converted into their trimethylsilyl ethers by reaction with N,O-bis-(trimethylsilyl)trifluoroacetamide (85°C, 60 min). Samples were dissolved in toluene and analyzed by GC-MS with a Trace GC gas chromatograph coupled to a quadrupole mass analyzer Trace MS (Thermo, Manchester, United Kingdom). The GC program and MS operation conditions were the same as described previously (29).
Data analysis.
The significance of the differences in MICs and ergosterol contents was determined by Student's t test (unpaired, unequal variance). A P value of <0.01 was considered significant. Statistical analysis was done with the SPSS package (version 13.0; SPSS S.L., Madrid, Spain).
Accession numbers for deduced proteins used in this work.
GenBank accession numbers for deduced proteins used in this work are as follows: Aspergillus nidulans I, EAA59846; A. nidulans II, EAA57846; Neurospora crassa I, EAA33687; N. crassa II, EAA33687; Magnaportha grissea, EAA46489; Leptosphaeria maculans, AAN27998; Ustilago maydis, EAK84444; S. cerevisiae, P32353; C. glabrata, AAB02330; C. albicans, AAC99343; C. dubliniensis, CAD13131; Schizosaccharomyces pombe I, BAA21457; S. pombe II, CAA22610; and Homo sapiens, NP008849.
Nucleotide sequence accession numbers.
The full nucleotide sequences of the erg3A, erg3B, and erg3C genes from A. fumigatus determined in this work appear in the GenBank nucleotide sequence database under accession numbers AY616449, AY616450, and AY616451, respectively.
RESULTS
Identification of putative A. fumigatus C-5 sterol desaturase (erg3) sequences.
The af51C7 sequence tag from an A. fumigatus strain was used to search the A. fumigatus genome databank at TIGR. The submitted sequence showed high percentages of nucleotide identity with three different DNA contigs (contig 790, 98%; contig 846, 67%; and contig 363, 59%). The three contigs, including 1 kb of adjacent sequence on each end, were pulled out using the TIGR facilities. Restriction maps and the deduced protein sequence from each of the three genes were obtained. Within the 1,134-bp sequence of A. fumigatus erg3A, there was a deduced open reading frame (ORF) of 335 amino acids. The ORF is interrupted twice by introns, based on the presence of matching consensus splice junctions (18). The sequence of erg3B (1,132 bp) can be deduced in an ORF encoding 352 amino acids, interrupted by a single intron (18). The 1,014-bp sequence of A. fumigatus erg3C is interrupted twice by intron sequences (18) and can be deduced in an ORF encoding 300 amino acids. The deduced amino acid sequences of the A. fumigatus gene fragments were used to carry out a BLASTp sequence similarity search in the Swissprot database of GenBank to identify homologous proteins. The results showed that Erg3A and Erg3B have a variable percentage of identity (from 46% to 89%) compared to a variety of filamentous fungi, yeasts, and human C-5 sterol desaturases. The Erg3C showed enough homology to belong to the Erg3 protein family, although percentages were much lower (36 to 61%). We therefore continued with the analysis of Erg3A and Erg3B.
Genomic organization and gene expression.
A. fumigatus genomic DNAs digested with different restriction enzymes were hybridized with fragments of each gene (erg3A and erg3B) used as probes. Southern hybridization confirmed that each probe hybridized to a different single band in each case, showing that each gene is present as a single copy in the genome of A. fumigatus (data not shown).
RT-PCR showed that both genes were expressed during hyphal growth in submerged culture. RT-dependent products of the expected sizes were amplified for each of the erg3 genes. Sequencing of each amplified band confirmed the presence of two introns in erg3A and one in erg3B.
A. fumigatus erg3A and erg3B full gene sequences and protein analysis.
Specific primers were used to amplify the full genomic sequence for each gene (erg3A and erg3B) from the A. fumigatus CM-237 strain (Table 1). erg3A was amplified using primers ERG321 and ERG322, and this product was sequenced with primers ERG3C, ERG3D, ERG3F, and ERG3Z in order to obtain the full sequence on both strands. To amplify erg3B, primers ERG3B3 and ERG3B4 were used and the product was fully sequenced with primers ERG3B1, ERG3B5, ERG3B9, and ERG3B10. The inferred 335-amino-acid protein Erg3A of A. fumigatus was compared to the other known complete Erg3 amino acid sequences obtained from GenBank. Strong homology was found with other Erg3 proteins from filamentous fungi: A. nidulans (II) Erg3 (60%); N. crassa (I) Erg3 (57%), and A.fumigatus deduced protein Erg3B (56%). Percentages of identity with the other yeast proteins were also very high (C. albicans, 46%; C. dubliniensis, 47%; S. cerevisiae, 46%; and C. glabrata, 47%). The inferred 352-amino-acid protein of erg3B was also compared to the same Erg3 sequences. A high degree of homology was shown with A. nidulans (II) Erg3 (89%), N. crassa (I) Erg3 (62%), and M. grissea Erg3 (60%). The homologies with yeast proteins were as follows: C. albicans, 40%; C. dubliniensis, 46%; C. glabrata, 47%; and S. cerevisiae, 47%.
Generation of erg3A− and erg3B− single mutant and erg3A− erg3B− double mutant strains.
We analyzed the functions of erg3A and erg3B by replacing the chromosomal copies of the genes with mutant versions in which the coding regions had been interrupted by the insertion of drug resistance markers. A hygromycin resistance marker was used for selection of single mutants (erg3A− and erg3B−), and a phleomycin resistance marker was used to allow for selection of double mutants (erg3A− erg3B−). To generate erg3A− mutants, spores of A. fumigatus CM-237 were electroporated using a 3.0-kb linear fragment released from pUM208 by NotI digestion (Fig. 2I). One hundred eighteen hygromycin-resistant transformants were screened by restriction enzyme digestion of genomic DNA and Southern hybridization. One of the transformants tested (CM-A80) appeared to have undergone gene replacement as shown by the shift of a hybridizing DNA fragment in strain CM-237 from 3.0 kb to 4.4 kb (Fig. 2I). Two more enzymes were used for gene replacement confirmation (data not shown).
To produce erg3B− mutants, spores of A. fumigatus CM-237 were electroporated using a 3.5-kb linear fragment released from pUM232 by ApaI/SacI double digestion (Fig. 2II). Three hundred sixty-six hygromycin-resistant transformants were screened. Any gene replacement event would be seen by a shift of a hybridizing DNA fragment in strain CM-237 from 4.0 kb to 5.4 kb. One transformant showed a band of 9.0 kb (with disappearance of the 4.0-kb wild-type band), suggesting a disruption of the gene with a tandem integration (Fig. 2II). Two more enzymes were used for confirmation of that specific integration (data not shown), and this was also verified by PCR using primers outside the replaced sequences.
For the construction of the erg3A− erg3B− double knockout mutant, spores of the A. fumigatus erg3A− mutant strain CM-A80 were electroporated using a 4.4-kb linear fragment released from pUM235 (containing the erg3B sequence interrupted by the phleomycin resistance marker) by ApaI/SacI double digestion (Fig. 2III). A total of 125 phleomycin-resistant transformants were analyzed by restriction enzyme digestion of genomic DNA and Southern hybridization. One transformant (CM-AB118) appeared to have undergone gene replacement of the interrupted erg3B gene, as shown by a shift of a hybridizing DNA fragment in strain CM-A80 from 4.0 kb to two different bands of 7.3 kb and 3.2 kb. As had occurred before, integration of the disruption vector also took place in tandem, including the insertion of one copy of the plasmid (Fig. 2III). Further analysis confirmed that the mutant was hygromycin resistant, and PCR amplification showed that CM-AB118 had retained the genotype of erg3A− disruption.
Morphology of mutant strains.
Spores (103) of wild-type strain CM-237 and mutant strains were plated in different media to study colony morphology and rate of growth for 72 h (38). No differences were found between the CM-A80, CM-B866, and CM-AB118 mutant strains and strain CM-237. Microscopically, no differences were observed in terms of hyphal or conidial morphology in any of the strains.
Antifungal susceptibility testing.
The results of antifungal susceptibility testing are shown in Table 2. The antifungal drug susceptibility profile of the mutants compared with the parental strain (CM-237) showed that there were no differences in susceptibility for any of the antifungal drugs tested. Results obtained by Etest also did not show any significant differences (data not shown).
TABLE 2.
Susceptibility testing of A. fumigatus wild type and mutants
| Strain | Genotype | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|---|
| AmB | Itraconazole | Voriconazole | Fluconazole | Ketoconazole | ||
| CM-237 | Wild type | 0.25-0.5 | 0.25-0.5 | 0.5 | >640 | 4-8 |
| CM-A80 | erg3A knockout | 0.25-0.5 | 0.12-0.5 | 0.5 | >640 | 4-8 |
| CM-B866 | erg3B knockout | 0.25-0.5 | 0.12 | 0.12-0.5 | >640 | 4-8 |
| CM-AB118 | erg3A erg3B knockout | 0.25-0.5 | 0.12-0.25 | 0.25 | >640 | 4-8 |
Testing was performed three times on different days.
Sterol composition.
The total ergosterol content was analyzed by high-pressure liquid chromatography, showing that this was similar for the wild-type strain CM-237 and the erg3A− mutant (5.99 ± 0.86 μg/mg and 6.58 ± 0.29 μg/mg, respectively). However, significant differences in total ergosterol were found for strains CM-B866 (2.02 ± 0.74 μg/mg) and CM-AB118 (1.75 ± 0.75 μg/mg). Moreover, the ergosterol UV spectrophotometric profiles for CM-B866 and CM-AB118 did not match with that of the commercial ergosterol, suggesting that other sterols could have accumulated as a result of erg3B deletion (data not shown). The wild type and mutants obtained during this study were consequently subjected to GC-MS analysis for the identification of sterol derivatives. The sterols were identified by comparison with known sterol spectra, and the results were expressed as percent area. As summarized in Table 3, ergosterol was the major sterol in the wild-type (CM-237) (73%) and CM-A80 (63%) strains. However, strains CM-866 and CM-AB118 showed a remarkable increase in three sterol fractions: 24-methylcholesta-7,22-dien-3β-ol (39% and 35%, respectively), 24-methylcholesta-7,22,24(28)-trien-3β-ol (19%), and 24-methylcholesta-7,24(28)-dien-3β-ol (11% and 12%, respectively). The accumulation of non-C-5 desaturated sterols in CM-B866 and CM-AB118 strain was coupled with a decrease of total ergosterol (11% and 17%, respectively).
TABLE 3.
Relative compositions of sterols identified in A. fumigatus control strain CM-237 and in the CM-A80, CM-B866, and CM-AB118 mutant strains
| Sterol | % in strain:
|
||||
|---|---|---|---|---|---|
| CM-237 | CM-A80 | CM-B866 | CM-AB118 | ||
| 24-Methylcholesta-5,7,9(11),22-tetraen-3β-ol | 1.07 | 3.27 | 1.20 | 0.84 | |
| 24-Methylcholesta-5,8,22-trien-3β-ol (lichesterol) | 0.89 | 0.84 | 0.06 | 0.10 | |
| 24-Methylcholesta-5,7,9,22-tetraen-3β-ol | 2.13 | 5.39 | 4.67 | 0.43 | |
| 24-Methylcholesta-5,7,22-trien-3β-ol (ergosterol) | 73.50 | 63.30 | 11.85 | 17.76 | |
| 24-Methylcholesta-7,22-dien-3β-ol | 2.12 | 1.08 | 39.14 | 35.73 | |
| 24-Methylcholesta-5,7,22,24(28)-tetraen-3β-ol | 0.44 | 0.48 | ND | ND | |
| 24-Methylcholesta-7,22,24(28)-trien-3β-ol | 0.40 | 0.88 | 19.76 | 19.95 | |
| 24-Methylcholesta-5,7,24(28)-trien-3β-ol | 1.49 | 1.14 | ND | ND | |
| 24-Methylcholesta-7,24(28)-dien-3β-ol (episterol) | 2.22 | 3.07 | 11.66 | 12.52 | |
| 24-Ethylcholest-5,7,22-trien-3β-ol | 4.65 | 4.93 | 1.48 | 0.79 | |
| 4,4,14-Trimethylcholesta-8,24-dien-3β-ol (lanosterol) | 2.19 | 2.43 | 2.06 | 1.79 | |
| 4α,24-Dimethylcholesta-8,24(28)-dien-3β-ol | 1.57 | 2.57 | 1.59 | 1.89 | |
| 4,4,14,24-Tetramethylcholesta-8,24(28)-dien-3β-ol (eburicol) | 2.72 | 3.69 | 2.32 | 2.91 | |
| 4,4,24-Trimethylcholesta-8,24(28)-dien-3b-ol | 4.60 | 6.90 | 4.20 | 5.29 | |
Boldface indicates major differences in percentages.
ND, not detected.
DISCUSSION
Ergosterol is the major sterol component in fungal membranes and contributes to a variety of cellular functions, including fluidity and integrity of the membranes and the proper function of membrane-bound enzymes (17). This fact makes enzymes involved in the ergosterol pathway attractive targets for antifungal agents. The C-5 sterol desaturase (Erg3) acts towards the end of the ergosterol biosynthetic pathway. It is located in the endoplasmic reticulum, where it catalyzes the introduction of a double bond between C-5 and C-6 in the B ring of the developing sterol molecule, converting episterol to 24-methylcholesta-5,7,24(28)-trien-3β-ol (45, 47) (Fig. 1).
erg3 genes have been isolated from many different yeasts and some filamentous fungi, due to their possible involvement in the study of resistance mechanisms of fungi against azole drugs and polyene compounds (1, 15, 26). The predicted amino acid sequences of A. fumigatus Erg3A and Erg3B revealed that they are different proteins and have a high degree of preservation among other C-5 sterol desaturase proteins. Both proteins have identity percentages that are sufficiently high for them to be considered members of the C-5 sterol desaturases family (34). Alignment between both A. fumigatus C-5 sterol desaturases and their orthologs in yeasts, filamentous fungi, plants, and humans showed that all have three conserved histidine domains. These domains are present in all known sequences of C-5 sterol desaturases and in a significant class of integral membrane enzymes that includes desaturases, hydroxylases, epoxidases, and acetylases (47). The preserved histidine-rich motifs are thought to contain the active site of the enzyme, being putative iron binding domains in C. albicans (32). Furthermore, all of them have the consensus motif for retention of transmembrane protein in the endoplasmic reticulum (21). The enzymatic conversion of lanosterol to ergosterol carries out enzymatic transformations that can occur in a different order in yeasts (14). A comparative study of the genes involved in Aspergillus ergosterol biosynthesis (erg) has shown the existence of several duplicated genes, including erg3 (13). The construction of strains with null mutations of genes involved in the biosynthesis of ergosterol and analysis of their sterol profiles could provide valuable information regarding the biosynthesis of ergosterol in A. fumigatus. We have identified different erg3 sequences that could encode C-5 sterol desaturases proteins in A. fumigatus. Despite the fact that three putative erg3 sequences had been identified, we started functional analysis with the Erg3 proteins that showed higher C-5 sterol desaturase homologies. As a result, erg3A, erg3B, and the combination of both were inactivated in order to study their roles in the A. fumigatus ergosterol biosynthesis pathway and their implication in antifungal drug susceptibility.
Functional analysis of Erg3A and Erg3B showed that neither is essential for cell viability and that the two genes are functionally quite different, despite their amino acid homology. Inactivation of the erg3B caused the accumulation of non-C-5 desaturated sterols: 24-methylcholesta-7,22-dien-3β-ol, 24-methylcholesta-7,22,24(28)-trien-3β-ol, and 24-methylcholesta-7,24(28)-dien-3β-ol. The lack of any obvious sterol composition alteration in the erg3A− strain implies that Erg3A might not be a C-5 sterol desaturase, although functional compensation by others enzymes (Erg3B and/or Erg3C) could not be ruled out. Alternatively, Erg3A could act in a different phase of the A. fumigatus cell cycle or under other growth conditions. To rule out the possibility that Erg3B could be compensating for the lack of Erg3A, we constructed a double mutant strain carrying disruption in genes encoding both proteins (Erg3A and Erg3B). The double mutant (erg3A− erg3B−) was indistinguishable from the erg3B− single mutant, including in their sterols profiles.
Despite the differences found in sterol composition between mutants and wild-type strains, no differences in AmB or azole drug susceptibility were detected. These results would dismiss any possibility of Erg3A and Erg3B being involved in the resistance mechanisms against antifungal drugs, at least until A. fumigatus erg3C− and erg3B− erg3C− mutants are analyzed. In yeast, targeted deletion of erg3 is always coupled with ergosterol depletion, although discrepancies with respect to AmB/azole susceptibility are seen between different species (20, 37, 43, 50). Since ergosterol is the target for AmB, high MICs to this antifungal would be expected in strains with decreased amounts of this sterol. The results obtained imply that AmB could be targeting other sterol derivatives in the cell, or, as other researchers have reported, that AmB standard antifungal susceptibility testing in A. fumigatus does not readily identify AmB resistance (22, 5). Moreover, in A. fumigatus deletion of the erg3 genes does not cause total ergosterol synthesis arrest, and this fact is consistent with the existence of more than one C-5 sterol desaturase enzyme that could compensate for the lack of one or even both Erg3 enzymes. Therefore, the involvement of Erg3C in the biosynthesis of the ergosterol pathway requires research. These experiments are under way in our laboratory.
Acknowledgments
This work was funded in part by grants MPY1120/03 from the Instituto de Salud Carlos III and SAF2002-02089 from the Ministry of Science and Technology. E. Mellado held a Ramón y Cajal contract from the Ministry of Science and Technology. L. Alcazar-Fuoli has a predoctoral fellowship from the Instituto de Salud Carlos III. Jordi F. Lopez's work is funded by an I3P contract from the Spanish Council for Scientific Research (CSIC).
We are grateful to J. P. Latge for providing the Erg3 initial sequence tag and helpful suggestions. Our thanks go to Fiona Westbury for correcting the English in the manuscript.
REFERENCES
- 1.Arthington, B. A., L. G. Bennett, P. L. Skatrud, C. J. Guynn, R. J. Barbuch, C. E. Ulbright, and M. Bard. 1991. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene 102:39-44. [DOI] [PubMed] [Google Scholar]
- 2.Arthington-Skaggs, B. A., D. W. Warnock, and C. J. Morrison. 2000. Quantitation of Candida albicans ergosterol content improves the correlation between in vitro antifungal susceptibility test results and in vivo outcome after fluconazole treatment in a murine model of invasive candidiasis. Antimicrob. Agents Chemother. 44:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bard, M., N. D. Lees, T. Turi, D. Craft, L. Cofrin, R. Barbuch, C. Koegel, and J. C. Loper. 1993. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 28:963-967. [DOI] [PubMed] [Google Scholar]
- 4.Chryssanthou, E. 1997. In vitro susceptibility of respiratory isolates of Aspergillus species to itraconazole and amphotericin B acquired resistance to itraconazole. Scand. J. Infect. Dis. 29:509-512. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2005. Reference method for broth dilution antifungal susceptibility. Testing of filamentous fungi. Approved standard document M38-A. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 6.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51-56. [DOI] [PubMed] [Google Scholar]
- 7.Cuenca-Estrella, M., T. M. Diaz-Guerra, E. Mellado, and J. L. Rodriguez-Tudela. 2001. Influence of glucose supplementation and inoculum size on growth kinetics and antifungal susceptibility testing of Candida spp. J. Clin. Microbiol. 39:525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, D., S. A. Leong, L. J. Wilson, and D. J. Henner. 1987. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene 57:21-26. [DOI] [PubMed] [Google Scholar]
- 9.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 10.Dannaoui, E., E. Borel, F. Persat, M. F. Monier, M. A. Piens, et al. 1999. In-vivo itraconazole resistance of Aspergillus fumigatus in systemic murine aspergillosis. J. Med. Microbiol. 48:1087-1093. [DOI] [PubMed] [Google Scholar]
- 11.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira, M. E., A. L. Colombo, I. Paulsen, Q. Ren, J. Wortman, J. Huang, M. H. Goldman, and G. H. Goldman. 2005. The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus. Med. Mycol. 43(Suppl. 1):S313-S319. [DOI] [PubMed] [Google Scholar]
- 14.Fryberg, M., A. C. Oehlschlager, and A. M. Unrau. 1973. Biosynthesis of ergosterol in yeast. Evidence for multiple pathways. J. Am. Chem. Soc. 95:5747-5757. [DOI] [PubMed] [Google Scholar]
- 15.Geber, A., C. A. Hitchcock, J. E. Swartz, F. S. Pullen, K. E. Marsden, K. J. Kwon-Chung, and J. E. Bennett. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Lopez, A., G. Garcia-Effron, E. Mellado, A. Monzon, J. L. Rodriguez-Tudela, and M. Cuenca-Estrella. 2003. In vitro activities of three licensed antifungal agents against Spanish clinical isolates of Aspergillus spp. Antimicrob. Agents Chemother. 47:3085-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooday, G. W. 1995. Cell membrane, p. 62-74. In N. A. R. Gow and G. M. Gadd (ed.), The growing fungus. Chapman & Hall, London, United Kingdom.
- 18.Gurr, S. J., S. E. Unkles, and J. R. Kinghorn. 1987. The structure and organization of nuclear genes of filamentous fungi, p. 93-139. In J. R. Kinghorn (ed.), Gene structure in eukaryotic microbes. IRL Press, Oxford, United Kingdom.
- 19.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, C. J., D. C. Lamb, N. J. Manning, D. E. Kelly, and S. L. Kelly. 2003. Mutations in Saccharomyces cerevisiae sterol C5-desaturase conferring resistance to the CYP51 inhibitor fluconazole. Biochem. Biophys. Res. Commun. 309:999-1004. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, E. M., K. L. Oakley, S. A. Radford, C. B. Moore, P. Warn, D. W. Warnock, and D. W. Denning. 2000. Lack of correlation of in vitro amphotericin B susceptibility testing with outcome in a murine model of Aspergillus infection. J. Antimicrob. Chemother. 45:85-93. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 25.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 26.Kontoyiannis, D. P., and R. E. Lewis. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135-1144. [DOI] [PubMed] [Google Scholar]
- 27.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann, P. A., R. M. Parmegiani, S. Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejanelle, L., J. F. Lopez, N. Gunde-Cimerman, and J. O. Grimalt. 2001. Ergosterol biosynthesis in novel melanized fungi from hypersaline environments. J. Lipid Res. 42:352-358. [PubMed] [Google Scholar]
- 30.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyazaki, Y., A. Geber, H. Miyazaki, D. Falconer, T. Parkinson, C. Hitchcock, B. Grimberg, K. Nyswaner, and J. E. Bennett. 1999. Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene 236:43-51. [DOI] [PubMed] [Google Scholar]
- 33.Mosquera, J., and D. W. Denning. 2002. Azole cross-resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagel, S. B. 2003. Function prediction from protein sequence, p. 65-79 In C. Orengo, D. Jones, and J. Thornton (ed.), Bioinformatics. Genes, proteins and computers. Bios Scientific Publishers, Oxford, United Kingdom.
- 35.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrikkou, E., J. L. Rodriguez-Tudela, M. Cuenca-Estrella, A. Gomez, A. Molleja, and E. Mellado. 2001. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J. Clin. Microbiol. 39:1345-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinjon, E., G. P. Moran, C. J. Jackson, S. L. Kelly, D. Sanglard, D. C. Coleman, and D. J. Sullivan. 2003. Molecular mechanisms of itraconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 47:2424-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeslev, M., and A. Kloller. 1995. Comparison of biomass dry weights and radial growth rates of fungal colonies on media solidified with different gelling compounds. Appl. Environ. Microbiol. 61:4236-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Tudela, J. L., E. Chryssanthou, E. Petrikkou, J. Mosquera, D. W. Denning, and M. Cuenca-Estrella. 2003. Interlaboratory evaluation of hematocytometer method of inoculum preparation for testing antifungal susceptibilities of filamentous fungi. J. Clin. Microbiol. 41:5236-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 1.21-1.32. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sanchez, O., and J. Aguirre. 2005. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet. Newslett. 43:48-51. [Google Scholar]
- 43.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slaven, J. W., M. J. Anderson, D. Sanglard, G. K. Dixon, J. Bille, I. S. Roberts, and D. W. Denning. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 45.Sugawara, T., Y. Fujimoto, and T. Ishibashi. 2001. Molecular cloning and structural analysis of human sterol C5 desaturase. Biochim. Biophys. Acta 1533:277-284. [DOI] [PubMed] [Google Scholar]
- 46.Tang, C. M., J. Cohen, and D. W. Holden. 1992. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 6:1663-1671. [DOI] [PubMed] [Google Scholar]
- 47.Taton, M., T. Husselstein, P. Benveniste, and A. Rahier. 2000. Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase studied by site-directed mutagenesis. Biochemistry 39:701-711. [DOI] [PubMed] [Google Scholar]
- 48.Vallejo, A. N., R. J. Pogulis, and L. R. Pease. 1995. Mutagenesis and synthesis of novel recombinant genes using PCR, p. 603-612. In G. S. Dveksler and C. W. Dieffenbach (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 50.Young, L. Y., C. M. Hull, and J. Heitman. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents Chemother. 47:2717-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]