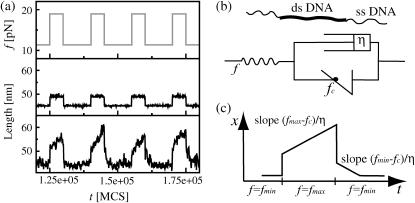
FIGURE 3.
Viscoelastic response of periodic DNA. (a) The shear force on an 80-bp dsDNA (with two 20-bp ssDNA linkers) is switched periodically between fmin = 11.4 pN and fmax = 19 pN (upper panel). The center and bottom panels show the extension-time-trace for heterogeneous and periodic DNA, respectively (energy parameters: ɛb = 1.11 kBT and ɛℓ = 2.8 kBT, roughly corresponding to AT basepairs at 50°C (18)). The time units are Monte Carlo steps, the real-time equivalent of which is discussed in Kinetic Rates (see article). The heterogeneous DNA responds only elastically to the force jumps, mostly due to stretching of the linkers. The length of the periodic DNA shows a similar elastic strain, but in addition, the molecule elongates at a finite speed due to sliding, since fmax > fc = 16.3 pN. When the molecule is relaxed, we find an elastic response and inward sliding, since fmin < fc. The length of the periodic DNA fluctuates strongly due to loop formation and annihilation. (b) The viscoelastic behavior displayed by a periodic DNA molecule can be described by a generalized Zener model, where harmonic springs are substituted by anharmonic elastic elements describing polymer elasticity and the restoring force fc. The ideal dashpot (with viscosity η) creates the viscous behavior of periodic dsDNA. (c) The response of the above idealized model to the same periodic force resembles the average response of periodic DNA.