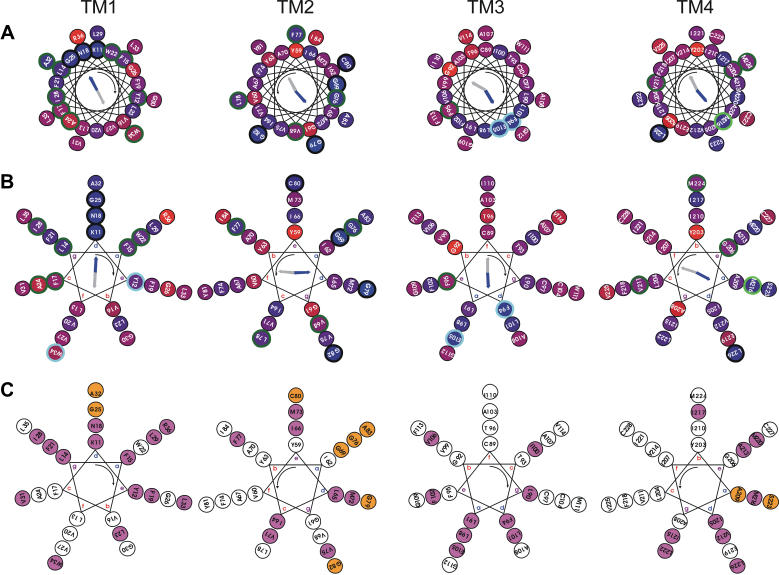
FIGURE 2.
Helical wheel representation of residue types and size conservation in CD81 transmembrane regions. (A) Representation of residue type conservation using a 3.6 residue-per-turn helical pitch (α-helix). The conservation of the residue position is indicated by the inner color of the circles in a linear red to blue scale with blue meaning 100% conserved. Residue positions that are conserved or homologous in all sequences are, respectively, circled in black and light blue. Residues positions that are variable among all sequences but conserved or homologous in all groups of sequences corresponding to a single tetraspanin species are, respectively, circled in dark and light green. The threshold for residue conservation or homology is 90%. Considered homologous residue families are: positive (H, K, R), negative-amidated (D, E, N, Q), hydroxylated (S, T), and aromatic (F, Y, W). The calculated conservation and hydrophobic moments are indicated as blue and gray arrows, respectively. (B) Same as panel A but using a 3.5 residue-per-turn helical pitch (coiled coil). (C) Representation of residue size conservation using a 3.5 residue-per-turn helical pitch (coiled coil). Residue positions that are conservatively occupied by a bulky (I, L M, V, W, Y, F, E, Q, N, K, R) and a small (G, A, S, T, C) side chain are, respectively, filled in magenta and orange. The threshold for size conservation is 95%. For calculation of conservation properties of TM1-4, local alignments of 81, 81, 88, and 64 sequences having 35, 35, 35, and 25% local identity with human CD81 were, respectively, used for each TM region. All helices are viewed from the intracellular side and the helix rotation sense is indicated by the curved arrow. For coiled coils (3.5 residues per turn) the canonical residue positions a–d are indicated.