TABLE 2.
Ligand-surrounding interaction energies from the MD simulations of P1 variants of OMTKY3 free in solution and when bound to elastase
| Elastase-OMTKY3
|
OMTKY3
|
|||||
|---|---|---|---|---|---|---|
| P1 residue |  |
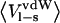 |
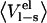 |
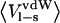 |
 * *
|
 † †
|
| Gly | −69.7 | −5.7 | −70.1 | −3.2 | −10.3 | −9.8 |
| Ala | −70.1 | −9.5 | −69.3 | −4.9 | −12.0 | −12.1 |
| Ser | −82.3 | −9.3 | −84.3 | −3.6 | −11.5 | −10.1 |
| Cys | −78.6 | −11.3 | −77.4 | −5.7 | −12.7 | −13.2 |
| Val | −67.4 | −16.9 | −69.6 | −8.9 | −12.6 | −13.6 |
| Thr | −77.3 | −14.9 | −83.0 | −5.6 | −12.1 | −12.2 |
| Leu | −71.1 | −16.9 | −71.3 | −10.7 | −12.4 | −13.1 |
| Ile | −68.3 | −20.0 | −70.4 | −10.6 | −13.4 | −13.8 |
| Met | −72.2 | −19.0 | −78.0 | −10.2 | −11.5 | −11.9 |
| Asn | −75.7 | −18.0 | −91.8 | −6.5 | −8.6 | −8.0 |
| Asp0‡ | −77.2 | −15.3 | −85.6 | −6.3 | −5.2 | −5.6 |
| Lys0‡ | −74.9 | −19.3 | −83.3 | −9.8 | −7.6 | −7.5 |
| Gln | −82.6 | −18.5 | −92.7 | −8.2 | −10.5 | −9.9 |
| Glu0‡ | −78.3 | −18.3 | −86.3 | −7.9 | −6.8 | −6.2 |
| His | −72.0 | −22.0 | −94.5 | −8.9 | −6.7 | −6.8 |
| Phe | −69.5 | −23.4 | −74.9 | −13.6 | −12.3 | −8.0 |
| Arg0‡ | −85.4 | −20.5 | −104.7 | −7.8 | −2.4 | −6.1 |
| Tyr | ND | ND | ND | ND | ND | −6.7 |
| Trp | −71.2 | −27.2 | −83.6 | −15.8 | −10.2 | −5.7 |
Values are given in kcal/mol.
Energies are calculated according to  and the error in these values is <1.0 kcal/mol. The coefficients used are from Model A in Table 4.
and the error in these values is <1.0 kcal/mol. The coefficients used are from Model A in Table 4.
Experimental binding data from Lu et al. (38).
0 indicates neutral side chain.  has been corrected with the free energy required to protonate/deprotonate the ionic side chain according to
has been corrected with the free energy required to protonate/deprotonate the ionic side chain according to  where pH is 8.3 (corresponding to the pH used in the association measurements) and pKa is 12.5, 10.7, 4.5, and 4.0 for Arg(5.7 kcal/mol), Lys(3.2 kcal/mol), Glu(5.1 kcal/mol), and Asp(5.8 kcal/mol), respectively.
where pH is 8.3 (corresponding to the pH used in the association measurements) and pKa is 12.5, 10.7, 4.5, and 4.0 for Arg(5.7 kcal/mol), Lys(3.2 kcal/mol), Glu(5.1 kcal/mol), and Asp(5.8 kcal/mol), respectively.
