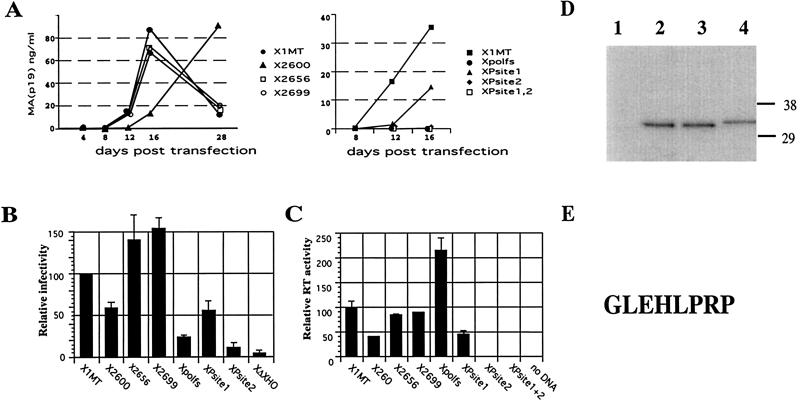
FIG. 4.
Infectivity and reverse transcriptase activity of pX1MT derivatives with mutations in TFP. (A) Spreading infection of virus produced after transfection of the indicated constructs into B5 cells determined by measuring MA (p19) resulting from ELISA of supernatants. Results for stop codon mutants (left panel) and protease cleavage site mutants and pX1MTpolfs (right panel) are compared to those for pX1MT. The experiments were repeated three times with similar results. (B) Single-cycle infection assays. Full-length pX1MT and mutant derivatives were used to package a luciferase gene-carrying HTLV-1 retroviral vector following transfection of the two constructs into 293 cells. Two independent transfections were carried out for each construct in each experiment. Supernatants from the transfected cells were standardized relative to the p19 concentration and used to infect 293 T cells. Results for pX1MT were set as 100, and values obtained for the other constructs are shown relative to that value. X1MTΔXho is a negative control with a deletion in env. Experiments whose results are shown in panels A and B were carried out as previously described (6). (C) Reverse transcriptase activities associated with wild-type and mutant viruses. 293 cells were transfected with the indicated constructs, and reverse transcriptase activity was determined directly for 4- and 20-μl volumes of supernatant by using the HS sensitivity RT kit from Cavidi (Upsalla, Sweden) in accordance with the manufacturer's directions. The reverse transcriptase activity per nanogram of p19 in the supernatant, determined by p19 ELISA, was calculated, and the activity of pX1MT was set as 100. The reverse transcriptase activities of the other constructs are given relative to that value. Results are derived from at least three independent transfection experiments. (D) Immunoblot of viral proteins from pCMVHT (6)-derived constructs expressing a truncated HA-tagged RT (Fig. 1A) with either no mutation (lane 2), a Psite1 mutation (lane 3), or a Psite2 mutation (lane 4). Lane 1 was loaded with the pelleted supernatant from a mock transfection. (E) The sequence derived for the amino terminus of the HA-tagged RT peptide. Viral proteins produced by CMVHT-polfs-polHA, which has the U3 region of the 5′ LTR replaced by the CMV immediate-early promoter and an insertion of a nucleotide into the second frameshift site, were affinity purified on anti-HA-agarose (Roche). Bound proteins were separated on NUPAGE gels, followed by transfer to an Immobilon membrane and staining with Coomassie brilliant blue. N-terminal sequence determination by Edman degradation of protein in the candidate band was carried out in an Applied Biosystems model 494CLC Procise sequenator. Phenylthiohydantoin derivatives of amino acids were catalyzed online with an Applied Biosystems model 785A/140C/610A analyzer.