FIG. 2.
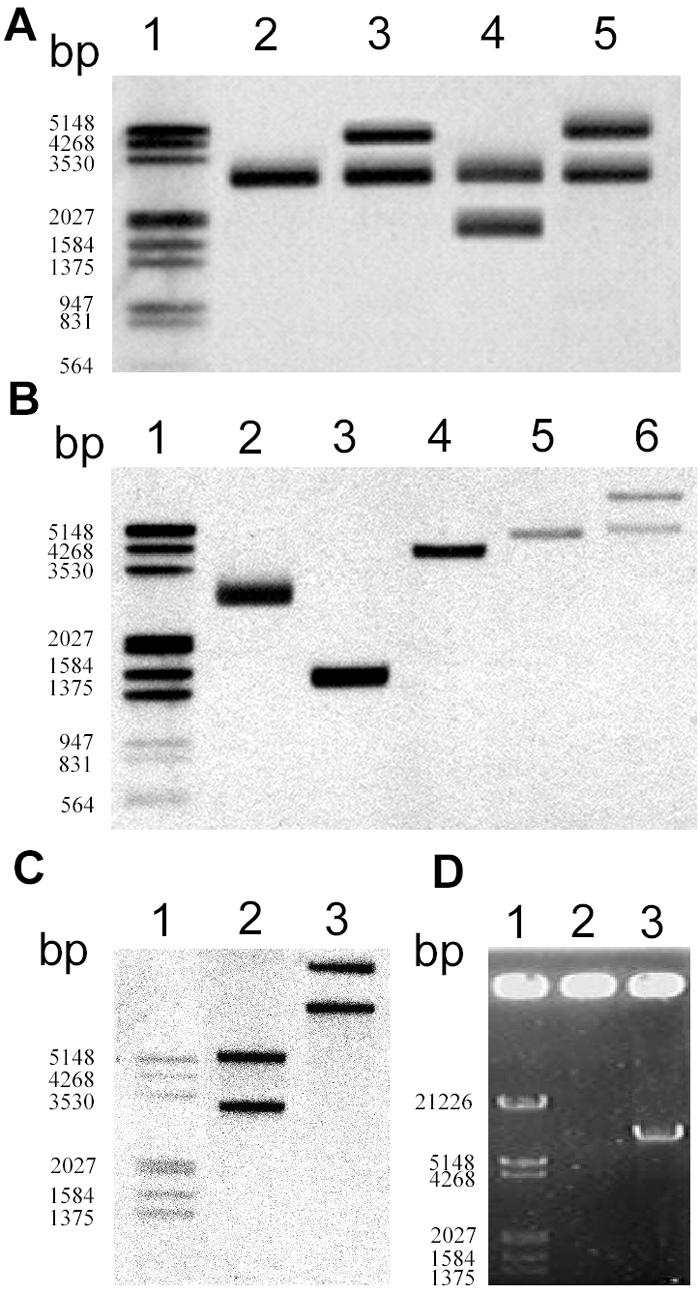
spnTIR region is mobile in S. marcescens SS-1. (A) Attempt to knock out spnT in SS-1 that led to the discovery of the duplication of the spnTIR region. Southern blot hybridization using tnpR as the probe after PstI digestion was performed to determine the restriction patterns of the chromosomal spnTIR region in mutants with potential spnT deletions. Lane 2, SS-1; lane 3, SS-1 KOT101; lane 4, SS-1 KOT102; lane 5, SS-1 KOT103. (B and C) Duplication of spnTIR in S. marcescens Jun-1. Chromosomal DNA of S. marcescens Jun-1 was digested by different restriction enzymes and probed with tnpR (B) or spnR (C). (B) Lane 2, PstI; lane 3, EcoRI; lane 4, NsiI; lane 5, EcoRV; lane 6, BamHI. (C) Lane 2, EcoRV; lane 3, BamHI. (D) Amplification of the potential transposon by single-primer PCR using S. marcescens SS-1 chromosomal DNA as the template and either TnP1 (lane 2) or TnP2 (lane 3) as the primer. Lane 1 contained a DNA marker. The numbers on the left indicate the sizes of markers.
