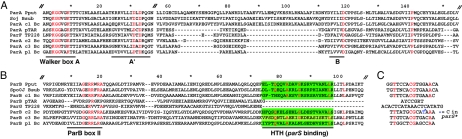
FIG. 2.
Parts of the aligned ParA (A) and ParB (B) amino acid sequences that distinguish subgroups. Residues that are identical (A) or nearly so (B) in all examples shown are in red. Numbers above show distances, not coordinates, and double hatch marks denote sequences not shown (18 residues just after the A box in A). The Walker A (nucleotide binding), A′ (catalytic), and B (Mg binding) motifs constitute the ATPase active site. ParBII is a motif identified previously (52). The helix-turn-helix (HTH) motifs are shaded green. The HTH of c3 is weakly predicted relative to the others. Note the short A′-B-box interval in the c2, c3, and p1 ParAs, very similar to the pTAR and TP228 homologues, and the absence of all HTH region homology in the latter ParBs. (C) Aligned parS and parS-like sequences with fully conserved bases in red. Note the dissimilarity of the pTAR and TP228 par region repeats.