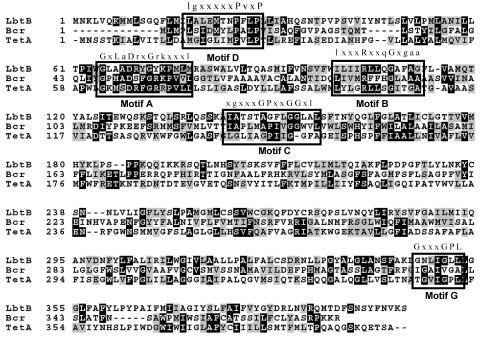
FIG. 6.
Amino acid sequence alignments of L. pneumophila LbtB with E. coli Bcr and TetA. The consensus sequences for conserved motifs A, B, C, D, G are labeled above the boxed-in areas; x is any amino acid, upper case is a highly conserved amino acid, and lower case is a conserved, but variable amino acid. Motif A, conserved in both 12- and 14-TMS families, is located in the cytoplasmic loop between TMS 2 and TMS 3 and may be involved in substrate binding as well as opening and closing of the channel (54). Motif B, located in TMS 4, is predicted to be involved in proton transfer (55). Motif C, located in TMS 5, is implicated in the direction of transport and is only found in those transport proteins with efflux capacity (30, 55). The function of motif D, located in TMS 1 in 12- and 14- TMS families, has not been investigated. Motif G, located in TMS 11, is found only in 12-TMS families, although its function is unknown (55).