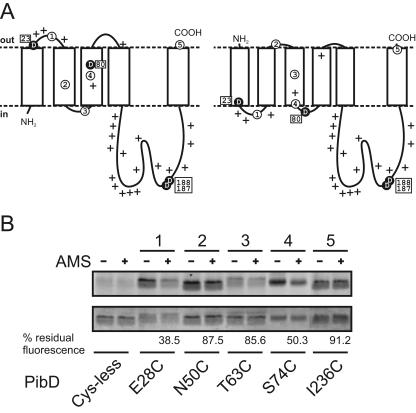
FIG. 3.
Membrane topology models of PibD and accessibility of single-cysteine mutants to AMS and fluorescein maleimide in everted membrane vesicles. (A) Representative membrane topology models predicted with TMHMM2.0 (left) and Phobius (right). Critical aspartic acid residues are highlighted as black circles, and positively charged residues (lysine and arginine) are marked with plus signs. Amino acid positions that were altered to cysteine are Glu28 (circled 1), Asn50 (circled 2), Thr63 (circled 3), Ser74 (circled 4), and Ile236 (circled 5). (B) Fluorescence labeling of PibD single-cysteine mutants. Everted E. coli vesicles without or with preincubation with AMS were solubilized, and protein was purified by immobilized metal affinity chromatography. Eluted protein was labeled with fluorescein maleimide and separated by SDS-PAGE. Fluorescent bands were visualized using an F1 Lumi-Imager (upper panel). Subsequently, the same gel was stained with Coomassie brilliant blue (lower panel). Numbers above lanes correspond to those used in panel A. Residual fluorescence of bands is expressed for each mutant as a ratio of AMS prelabeled samples to mock-treated samples after correction for protein loading and the background signal of the cysteineless mutant.