TABLE 1.
The DNA sequences used in the calculations
| Sequence | N (bp) | Experiment reference | −ΔH° (kcal/mol)* | −ΔS° (cal/mol.K)* | −ΔG°37 (kcal/mol)* |
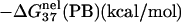 † †
|
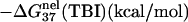 ‡ ‡
|
|---|---|---|---|---|---|---|---|
| GCAGC | 5 | – | 35.7 | 98.1 | 5.3 | 3.9 | 4.0 |
| GCATGC | 6 | (36) | 43.6 | 121.6 | 5.9 | 4.3 | 4.5 |
| CCAAACA | 7 | (78) | 46.8 | 130.8 | 6.2 | 4.2 | 4.5 |
| GGAATTCC | 8 | (79–81) | 55.2 | 156.0 | 6.8 | 4.5 | 4.8 |
| GCCAGTTAA | 9 | (37) | 63.1 | 174.8 | 8.9 | 6.3 | 6.7 |
| ATCGTCTGGA | 10 | (35,82) | 70.5 | 192.0 | 11.0 | 8.0 | 8.5 |
| CCATTGCTACC | 11 | (35) | 81.1 | 222.5 | 12.1 | 8.8 | 9.4 |
| CAAAGATTCCTC | 12 | (83) | 87.4 | 243.8 | 11.8 | 8.2 | 9.0 |
| AGAAAGAGAAGA | 12 | (84,85) | 83.1 | 231.2 | 11.4 | 7.8 | 8.6 |
| CCAAAGATTCCTC | 13 | – | 95.4 | 263.7 | 13.6 | 9.6 | 10.4 |
The thermodynamic data are calculated at standard state (1 M NaCl) through the nearest-neighbor model with the thermodynamic parameters of SantaLucia (22).
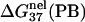 is computed from Eq. 3 with
is computed from Eq. 3 with 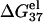 calculated from the Poisson-Boltzmann theory.
calculated from the Poisson-Boltzmann theory.
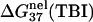 is calculated from Eq. 3 with
is calculated from Eq. 3 with 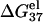 calculated through the tightly bound ion model. For each of the listed sequences, we have performed the thermodynamic calculations for both the NaCl and the MgCl2 solutions of different ion concentrations.
calculated through the tightly bound ion model. For each of the listed sequences, we have performed the thermodynamic calculations for both the NaCl and the MgCl2 solutions of different ion concentrations.
