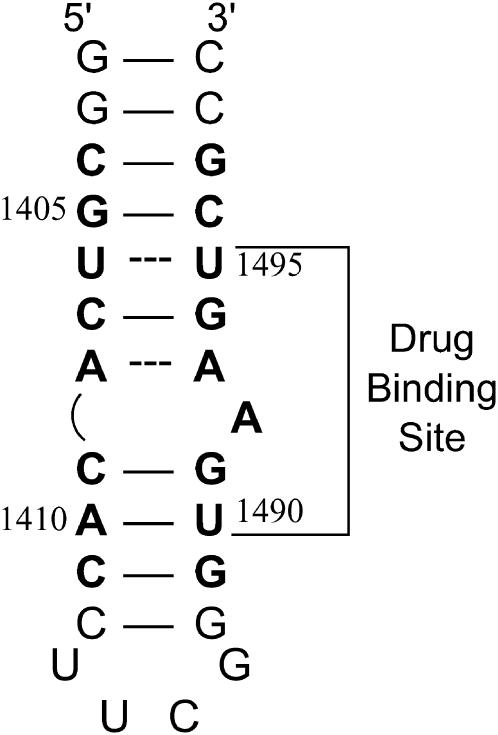
FIGURE 5.
NMR-derived (53–55) secondary structure of the E. coli 16 S rRNA A-site model oligonucleotide used in this study. Watson-Crick basepairs are denoted by solid lines, whereas mismatched basepairs are denoted by dashed lines. Bases present in E. coli 16 S rRNA are depicted in bold face, and are numbered as they are in 16 S rRNA. The paromomycin binding site, as revealed by NMR and footprinting studies (53,54,56), is indicated.