TABLE 2.
The model parameters*
| Rate constants | Other parameters | |
|---|---|---|
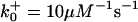 |
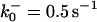 |
|
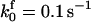 |
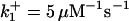 |
p = 4 |
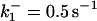 |
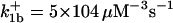 |
[p53thresh] = 0.004 μM |
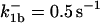 |
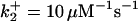 |
μ = 0.006 s−1 |
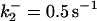 |
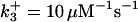 |
ΩApaf-1 = 3 × 10−4μM/s |
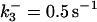 |
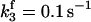 |
ΩIAP = 3 × 10−5μM/s |
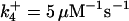 |
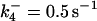 |
Ωpro3 = 3 × 10−4μM/s |
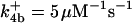 |
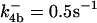 |
Ωpro9 = 3 × 10−4μM/s |
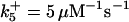 |
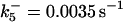 |
ΩBid = 3 × 10−5μM/s |
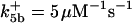 |
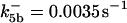 |
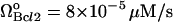 |
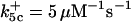 |
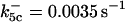 |
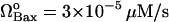 |
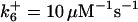 |
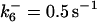 |
Ωcytmit = 3 × 10−4μM/s |
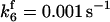 |
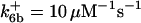 |
[p53] = 0.0066 μM |
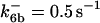 |
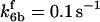 |
|
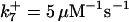 |
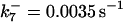 |
|
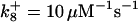 |
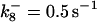 |
|
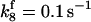 |
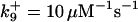 |
|
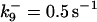 |
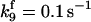 |
|
| k11 = 10 s−1 | k12a = 10 μM−1s−1 | |
| k12b = 10 μM−1s−1 | k13 = 10 μM−1s−1 | |
| k14 = 10 μM−1s−1 | ||
*All rate constants, except for 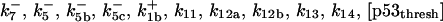 , are set up to within one order of magnitude difference with respect to those used by Asthagiri and Lauffenburger (43) in their simulations of mitogen-activated protein kinase pathways. The ratio of
, are set up to within one order of magnitude difference with respect to those used by Asthagiri and Lauffenburger (43) in their simulations of mitogen-activated protein kinase pathways. The ratio of 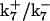 ensures an equilibrium constant of 0.7 nM (56), as well as those used for defining
ensures an equilibrium constant of 0.7 nM (56), as well as those used for defining 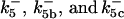 . The values of
. The values of 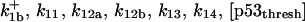 are suitably set to achieve fluxes comparable to other compounds' fluxes;
are suitably set to achieve fluxes comparable to other compounds' fluxes; 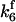 is assigned a value smaller than
is assigned a value smaller than 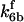 because of experimental observations by Riedl et al. (30). The degradation constant of all species is assigned as 0.006 s−1 unless specified otherwise in the text. This value is within the range of parameters used for degradation rate constants adopted by Chen et al. (42) The rates of synthesis are within the range of values used by Chen et al. (42).
because of experimental observations by Riedl et al. (30). The degradation constant of all species is assigned as 0.006 s−1 unless specified otherwise in the text. This value is within the range of parameters used for degradation rate constants adopted by Chen et al. (42) The rates of synthesis are within the range of values used by Chen et al. (42).
