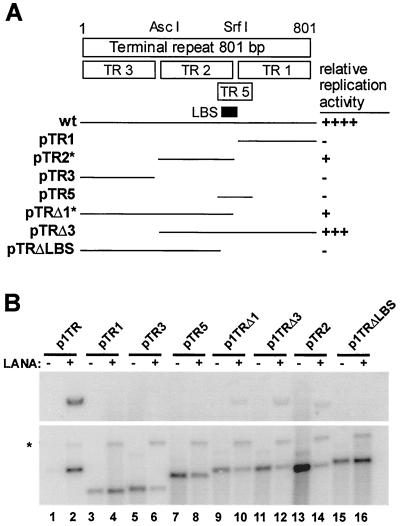
FIG. 2.
Mapping of cis requirements for LANA-dependent DNA replication. (A). Illustration of TR unit indicating constructs TR1, -2, -3, and -5, which have been used to identify the LBS within the TR. Below this, the solid lines represent TR-derived fragments that were inserted into either pCRII or pBS and were tested in short-term replication assays (pTR1, nt 1 to 268; pTR2, nt 268 to 581; pTR3, nt 581 to 801; pTR5, nt 551 to 675; pTRΔ1, nt 1 to 581; pTRΔ3, nt 268 to 801; and pTRΔLBS, nt 1 to 559). Also indicated are the relative replication efficiencies of these plasmids compared to wt p1TR, which were derived by comparing the ratios of input to replicated DNA after densitometric measurement. The asterisks at pTR2 and pTRΔ1 indicate that the LBS is not fully contained in these constructs. (B) Mapping data from short-term replication assays in 293 cells. All assays were performed as described in Fig. 1. Odd-numbered lanes indicate plasmids in the absence of LANA; even-numbered lanes indicate plasmids in the presence of LANA. Only wt p1TR, pTRΔ1, pTRΔ3, and pTR2 replicate, as shown in the upper blot in lanes 2, 10, 12, and 14; the lower panel shows linearized plasmids as a loading control. The asterisk shows the linearized pcDNA3/ORF73, since the blots were hybridized to a probe containing the entire pCRII/TR plasmid.