Abstract
Characterization of recent outbreaks of fatal encephalitis in southeast Asia identified the causative agent to be a previously unrecognized enveloped negative-strand RNA virus of the Paramyxoviridae family, Nipah virus. One feature linking Nipah virus to this family is a conserved cysteine-rich domain that is the hallmark of paramyxovirus V proteins. The V proteins of other paramyxovirus species have been linked with evasion of host cell interferon (IFN) signal transduction and subsequent antiviral responses by inducing proteasomal degradation of the IFN-responsive transcription factors, STAT1 or STAT2. Here we demonstrate that Nipah virus V protein escapes IFN by a distinct mechanism involving direct inhibition of STAT protein function. Nipah virus V protein differs from other paramyxovirus V proteins in its subcellular distribution but not in its ability to inhibit cellular IFN responses. Nipah virus V protein does not induce STAT degradation but instead inhibits IFN responses by forming high-molecular-weight complexes with both STAT1 and STAT2. We demonstrate that Nipah virus V protein accumulates in the cytoplasm by a Crm1-dependent mechanism, alters the STAT protein subcellular distribution in the steady state, and prevents IFN-stimulated STAT redistribution. Consistent with the formation of complexes, STAT protein tyrosine phosphorylation is inhibited in cells expressing the Nipah virus V protein. As a result, Nipah virus V protein efficiently prevents STAT1 and STAT2 nuclear translocation in response to IFN, inhibiting cellular responses to both IFN-α and IFN-γ.
Interferons (IFNs) are the primary innate antiviral cytokines in mammalian cells and also regulate aspects of the adaptive immune response (3, 5, 27). Both IFN-α/β and IFN-γ can induce an antiviral state in cells as an end point of signal transduction through the JAK-STAT pathway. The general paradigm for this signaling pathway involves IFN-dependent receptor-mediated tyrosine phosphorylation of latent cytoplasmic STAT proteins to produce DNA-binding oligomers that are competent for nuclear translocation. IFN-α/β responses are mediated primarily by an activated transcription complex, ISGF3, that consists of heterodimeric STAT1 and STAT2 in association with a DNA binding subunit, IRF9. ISGF3 binds to a DNA element, ISRE, in IFN-α/β target gene promoters, inducing their transcription. IFN-γ responses are also mediated by a STAT-containing transcription complex consisting of STAT1 homodimers. The STAT1 homodimer binds to a DNA element termed GAS to induce the transcription of a distinct but overlapping set of IFN-γ target genes. In both cases, these IFN-stimulated gene products produce a general antiviral state in the cell that inhibits the replication of diverse virus species.
The Paramyxoviridae family represents a wide variety of enveloped minus-strand RNA viruses including a number of established and emerging human and animal pathogens (12). Several paramyxoviruses, particularly those in the Rubulavirus genus, have been recently demonstrated to interfere with IFN-induced antiviral responses by targeting the STAT1 and/or STAT2 proteins for proteasomal degradation. For example, mumps virus and simian virus 5 (SV5) target STAT1 for degradation, while human parainfluenza virus type 2 (HPIV2) targets STAT2 (7, 11, 20).
The exact mechanisms underlying STAT protein targeting by the rubulaviruses are not completely understood, but it is clear that the anti-STAT effects are mediated by a single viral protein, termed V (7). The V protein is not related to any cellular proteins and does not appear to be a protease itself; instead, it acts in concert with cellular factors to mediate STAT degradation. For SV5-induced STAT1 degradation and HPIV2-induced STAT2 degradation, the targeting complex requires at least V, STAT1, and STAT2 to be present in the host cell (21). The role for a nontarget STAT protein in a STAT-targeting complex is not apparent, but available evidence indicates that the requirement for the confederate STAT protein is completely independent of IFN-α/β signal transduction or ISGF3 functions and can be supplied by a STAT protein N-terminal fragment (21). Moreover, the ability of SV5 to induce the degradation of STAT1 and to antagonize IFN signaling is species specific. IFN evasion by SV5 is very efficient in primate cells but restricted in murine cells (6). The cellular basis for the differential ability of murine and human cells to create an innate antiviral response to SV5 was recently uncovered (19). The failure of SV5 to antagonize IFN responses in the mouse system is not due to intrinsic defects in the SV5 proteolytic target, STAT1, since IFN responses that are dependent on murine STAT1 are efficiently blocked in otherwise human cells. Instead, it was found that differences between human and mouse STAT2 proteins provide the molecular basis for SV5 species specificity in IFN antagonism and STAT degradation, providing further support for the critical role of the nontarget STAT2 in V-dependent STAT1 degradation.
An outbreak of fatal encephalitis and zoonotic disease in southeast Asia between 1998 and 1999 that resulted in over 100 human deaths was subsequently linked to a previously unidentified paramyxovirus, named Nipah virus (4); reviewed comprehensively in reference 8 and citations therein. Nipah virus is closely related to another recently emerged fatal paramyxovirus, Hendra virus (28). These two viruses differ significantly from the other Paramyxovirus genera, and classification of these viruses in a new paramyxovirus genus, Henipavirus, has been proposed (28, 29). Since these viruses have only recently appeared as human pathogens, little is known about their life cycles or their ability to interact with host cells. Investigation of the mechanisms underlying immune evasion by these viruses and identification of the virus-encoded factors that contribute to their ability to rapidly disseminate in a variety of host species is essential for understanding their pathogenesis.
Because V protein-dependent IFN antagonism has been linked to successful paramyxovirus host range determination, replication, and pathogenesis, the Nipah virus V protein is a candidate pathogenesis-determining factor and was therefore tested for IFN inhibition capabilities. Like other paramyxovirus V proteins, expression of the Nipah virus V protein inactivates IFN signaling by direct inhibition of STAT1 and STAT2 function. However, the Nipah V protein differs from other paramyxovirus V proteins in its subcellular distribution and the mechanism used to inhibit cellular IFN responses. Results indicate that Nipah virus V protein accumulates in the cytoplasm as a result of a Crm1-dependent nuclear export mechanism. Nipah virus V protein also associates tightly with both STAT1 and STAT2. As a result, the STAT proteins are retained in the cytoplasm, preventing IFN-induced STAT activation and nuclear translocation, thereby inhibiting cellular responses to either IFN-α or IFN-γ.
MATERIALS & METHODS
Cell culture.
Human 2fTGH, 293T, and U6A cells (STAT2-deficient 2fTGH daughter cell line [13]) were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% Cosmic calf serum (Hyclone) and 1% penicillin-streptomycin (Gibco-BRL).
Plasmids, transfection, and reporter gene assays.
Nipah V cDNA in plasmid pTM1 was obtained from Paul Rota (Centers for Disease Control and Prevention, Atlanta, Ga.). PCR primers homologous to the 5′ and 3′ ends of the Nipah V open reading frame were used to amplify Nipah V cDNA and add BglII and NotI restriction sites. The PCR product was ligated to the-pEFN vector (gift of Netai Singh, Mount Sinai School of Medicine) with influenza virus hemagglutinin (HA) or FLAG octapeptide epitope tags. Nipah V plasmids were sequenced and determined to be identical to the GenBank Nipah V cDNA sequence (accession no. NC 002728). Epitope-tagged HPIV2 and SV5 V protein expression vectors were generated in a similar manner as described elsewhere (20, 21).
High-efficiency transient transfection of 293T cells was carried out on 60 to 80% confluent 60-mm dishes using standard calcium phosphate procedures (1). To estimate transfection efficiency (typically greater than 90% of cells transfected), 0.1 μg of the plasmid EGFP-N1 was cotransfected with each sample and visualized by inverted fluorescence microscopy prior to lysis.
For luciferase assays, 293T cells in 60-mm dishes were transfected with 2 μg of reporter gene (for IFN-α, 5× ISRE-luciferase; for IFN-γ, 4× M67 SIE-luciferase [21]), 6 μg of HA-tagged Nipah V plasmid, and 0.5 μg of cytomegalovirus (CMV)-LacZ. IFN-α (1,000 U/ml), IFN-γ (5 ng/ml), or leptomycin B (LMB) was added directly to the culture medium as indicated for the individual experiments. Data represent the average values of triplicate samples normalized to cotransfected β-galactosidase activity, expressed as a percentage of control IFN-stimulated sample.
Cell extracts, immunoblotting, and immunoprecipitation.
For preparation of cell extracts, samples in 60-mm dishes were washed once with ice-cold phosphate-buffered saline and subsequently lysed with 250 μl of whole-cell extract buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% IGEPAL, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol [DTT]) supplemented with protease inhibitor cocktail (Complete; Boehringer Mannheim) and sodium vanadate (0.1 mM). Lysates were incubated on ice for 10 min and centrifuged for 10 min at 20,800 × g at 4°C. Supernatants were analyzed on sodium dodecyl sulfate (SDS)—12% polyacrylamide gels. Proteins were then transferred to nitrocellulose, probed with antibodies for STAT1 (Upstate Biotechnology, no. 06-501), STAT2 (Santa Cruz, no. sc-476), phosphotyrosine 701-STAT1 (Cell Signaling Technologies, no. 9171), or FLAG epitope tag (Sigma-Aldrich), and visualized by chemiluminescence (NEN Life Sciences).
Cell extracts were immunoprecipitated using M2 FLAG affinity gel (Sigma-Aldrich). Cell extracts (700 μg) were incubated with 5 μl of M2 beads in a 50% slurry, rocked for 8 h at 4°C, and washed five times with 1 ml of whole-cell extract buffer. Samples were boiled in 30 μl of SDS gel loading buffer, and 15 μl was loaded directly onto an SDS-12% polyacrylamide gel for analysis.
Gel filtration chromatography
For gel filtration analysis, 293T cells were transfected with 10 μg of HA-tagged Nipah V expression vector or pEGFP-N1 plasmid. The cells were collected, centrifuged in 1 ml of PBS, and lysed in 500 μl of extract buffer (PBS, 0.5% IGEPAL, 1 mM DTT). Extracts were fractionated on a Superdex 200 Hiload 1660 fast protein liquid chromatography column (120-ml bed volume) in PBS-1 mM DTT with a flow rate of 0.5 ml/min. After 40 ml of void volume, 80 1-ml fractions were collected. Every third sample was lyophilized to 100 μl, mixed with 20 μl of SDS gel loading buffer, boiled for 5 min, and separated on SDS-12% polyacrylamide gels. After transfer to nitrocellulose, proteins were detected by immunoblotting. Molecular weight calibration was carried out with high- and low-molecular-weight calibration kits (Amersham-Pharmacia) as recommended by the manufacturer using the following standards: ferritin, 440,000; catalase, 232,000; aldolase, 158,000; albumin, 67,000; ovalbumin, 43,000; chymotrypsinogen A, 25,000; and RNase A, 13,700.
Indirect immunofluorescence.
For indirect-immunofluorescence experiments, 2FTGH and U6A cells were grown to 60 to 80% confluence on Permanox chamber slides (Nalgene Nunc) and transfected with empty vector or HA-V expression plasmids using Superfect reagent (Qiagen) as recommended by the manufacturer (typically achieving 1 to 10% transfection efficiency). LMB (10 ng/ml) was added 3 h prior to fixation; IFN treatment was for 30 min prior to fixation. At 24 h after transfection, the cells were fixed in 200 μl of 1% formaldehyde in PBS for 15 min and permeabilized in ice-cold 1:1 methanol-acetone for 10 min at −20°C. After five washes with PBS, samples were blocked with 1% bovine serum albumin in PBS for 15 min at 37°C. After every subsequent antibody exposure, samples were washed and blocked. Antibody staining was performed sequentially, with HA antibody at a 1:300 dilution (2.8 μg/ml) first. Texas Red conjugated to mouse immunoglobulin G was used to visualize HA-V proteins. The second stain for either STAT1 (0.02 μg/ml) or STAT2 (4 μg/ml) was detected with fluorescein isothiocyanate-conjugated rabbit immunoglobulin G. STAT1 and STAT2 polyclonal antisera used for immunofluorescence were precleared on fixed and permeabilized STAT1-deficient U3A cells or STAT2-deficient U6A cells to reduce nonspecific background staining. Images were obtained using a Leica TCSSP confocal microscope. Representative fields illustrating transfected and untransfected cells are illustrated, but the effects of Nipah virus V protein on STATs were observed with 100% penetrance, even in cells with relatively low V protein levels.
RESULTS
Expression of Nipah V antagonizes IFN-α and IFN-γ signaling.
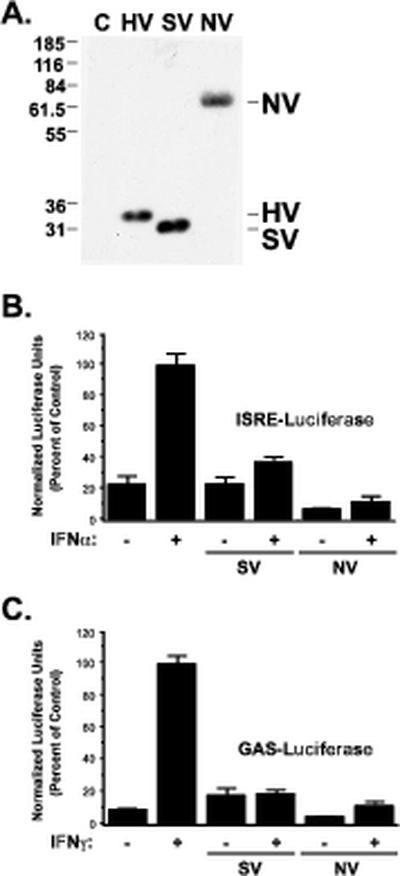
Database searching algorithms indicate that Nipah virus V protein has no obvious homology to any cellular protein. The Nipah virus V protein N terminus is unique compared to other paramyxovirus proteomes, but Nipah virus V protein has approximately 52% amino acid identity within the C-terminal cysteine-rich domain, a conserved Zn2+ binding domain that is essential for virus replication in IFN-competent systems (10) and for in vitro V protein activities including cell cycle arrest and protein interactions (14, 15, 22). To examine the potential of Nipah virus for IFN evasion, a cDNA encoding Nipah virus V protein was subcloned into a mammalian expression vector downstream of an epitope tag (influenza virus HA or FLAG epitope tag) and the vectors were used to transfect cultured human cells. Immunoblotting with tag-specific antiserum reveals a single species of Nipah virus V protein migrating with an apparent molecular mass of ∼70 kDa (larger than its predicted molecular mass of ∼50.3 kDa, possibly the result of posttranslational modifications), distinct in size from the HPIV2 V protein or SV5 V protein expressed in parallel (Fig. 1A).
FIG. 1.
Expression of the Nipah virus V protein inhibits IFN-α and IFN-γ signaling. (A) Human fibrosarcoma 2FTGH cells were transfected with empty vector (lane C) or expression vectors for HA-tagged SV5 V (lane SV), HPIV2 V (lane HV), or Nipah V (lane NV), and lysates were separated by SDS-PAGE (12% polyacrylamide). V proteins were visualized by immunoblotting with antiserum to the HA epitope tag. (B) 2FTGH cells were transfected with an ISRE-luciferase reporter gene and either empty vector, SV5 V (SV), or Nipah V (NV) expression vectors as indicated. The cells were treated with 1,000 U of IFN-α per ml 8 h prior to lysis and luciferase assays (+) or left untreated (−). (C) Same as in panel B but using an IFN-γ-responsive GAS-luciferase reporter gene and treatment with 5 ng/ml of IFN-γ (or no treatment). All bars represent average values from triplicate samples, normalized to cotransfected CMV-LacZ and expressed as percentages of IFN stimulated controls; error bars indicate standard deviations.
The biological activity of other paramyxovirus V proteins has been correlated with their ability to antagonize IFN signal transduction, thereby blocking ISGF3-dependent transcription. To test the potential of Nipah virus V protein to function as an IFN antagonist, IFN-α/β- and IFN-γ-responsive luciferase reporter gene assays were conducted in the presence and absence of expressed V proteins. IFN-α treatment resulted in induction of ISRE-luciferase reporter gene activity, reflecting the actions of the endogenous ISGF3 transcription complex (Fig. 1B). Expression of the Nipah virus V protein completely blocked IFN-α-responsive transcription, as well as or better than the control SV5 V protein. For IFN-γ signaling, a homodimer of STAT1 is the active transcription complex generated by receptor occupation. In reporter gene assays for IFN-γ signaling, similar Nipah virus V protein-dependent inhibition was observed (Fig. 1C). Together, these results reveal that Nipah virus V protein functions effectively as a potent inhibitor of both IFN-α and IFN-γ transcriptional responses, possibly through an inhibitory mechanism that attacks a component common to both IFN signaling pathways.
Nipah V protein binds STATs without degradation.
The mechanism of IFN evasion used by SV5 and HPIV2 is known to involve V-induced degradation of STAT proteins (7, 20). To determine if Nipah virus V protein shares this mechanism of IFN evasion, STAT degradation assays were performed. A comparison of the steady-state levels of STAT1 and STAT2 in control cells with those in SV5 V protein-expressing cells clearly demonstrates STAT1 degradation induced by SV5 V protein. In contrast, Nipah virus V protein-specific decreases in either the STAT1 or STAT2 protein levels were not observed (Fig. 2A), despite a high (≥90%) transfection efficiency as determined by a cotransfected green fluorescent protein (GFP) expression plasmid (data not shown). The efficient IFN antagonism induced by Nipah V protein expression in the absence of observed STAT1 or STAT2 degradation is consistent with an alternate mode of action for this V protein.
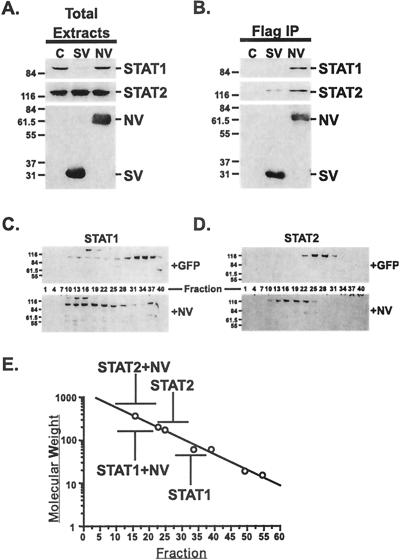
FIG. 2.
Nipah virus V binds to STAT1 and STAT2 without inducing STAT degradation. (A) 293T cells were transfected with empty vector (lane C), FLAG-tagged SV5 V (lane SV), or FLAG-tagged Nipah virus V (lane NV) expression vectors. Whole-cell extracts were prepared 36 h later, and 20 μg of protein was separated by SDS-PAGE (12% polyacrylamide) and processed for immunoblotting with antisera to STAT1, STAT2, or FLAG. Positions of prestained molecular weight standards (in thousands) and STAT1, STAT2, Nipah V protein (NV), and SV5 V protein (SV) are indicated. (B) Extracts as in panel A (700 μg of total protein) were immunoprecipitated with FLAG affinity gel (Sigma) and analyzed by immunoblotting. (C and D) Gel filtration analysis. Whole-cell extracts from control or NV-expressing cells were separated by chromatography on a Superdex-200 column. Positions of STAT1 (C) and STAT2 (D) were determined in control cells (+GFP) or in the presence of Nipah V protein (+NV) by immunoblotting of every third fraction. (E) Gel filtration data plotted in comparison to a standard curve. Elution positions are indicated for STAT1 and STAT2 in the presence and absence of Nipah V protein (NV) with respect to calibration standards (open circles).
Previous studies of IFN antagonism by SV5 and HPIV2 demonstrated that a multiprotein complex that includes V protein and both STAT1 and STAT2 proteins is essential for IFN antagonism (21). The ability of Nipah virus V protein to interact with STAT1 and STAT2 was tested in comparison with the STAT1-degrading SV5 V protein. Immunoprecipitation of FLAG-tagged SV5 V protein resulted in coprecipitation of a small amount of STAT2, in agreement with prior results (21) (Fig. 2B). The FLAG-Nipah virus V protein immunoprecipitation resulted in coprecipitation of STAT1 and STAT2, with higher yields of STAT1 and STAT2 coprecipitation with Nipah V protein than was observed for SV5 V-STAT2 coprecipitation, possibly reflecting a higher binding affinity.
The apparent high affinity of Nipah V protein for STAT1 and STAT2 indicated that gel filtration chromatography could be used to determine the extent of Nipah virus V protein-STAT complex formation and for estimating the size of the complex. The standard model for STAT protein activation involves IFN-induced conversion of latent monomeric STATs to activated oligomers, based primarily on size estimates from glycerol gradient sedimentation in unstimulated or IFN-treated whole-cell detergent extracts (25). More recently, high-molecular-weight heteromeric STAT1- and STAT3-containing complexes have been identified in detergent-free cytosolic extracts from unstimulated cells (reviewed in reference 24). The pattern of STAT1 elution from the gel filtration column was determined by immunoblotting for control 293T cells transfected only with a GFP expression vector. A small degree of heterogeneous STAT1 migration in the high-molecular-weight fractions was observed, but a substantial peak of STAT1 eluted in fractions 28 to 37, in agreement with the apparent size of STAT1 on SDS-polyacrylamide gel electrophoresis (PAGE) or in glycerol gradients of approximately 91 kDa (Fig. 2C). STAT2 eluted from the column in a single peak encompassing fractions 22 to 31, again in general agreement with the STAT2 apparent molecular mass in SDS-PAGE of approximately 113 kDa (9) (Fig. 2D). Expression of Nipah virus V protein in the cells induced a shift in the elution profiles of both STAT1 and STAT2 to a high-molecular-weight position encompassing fractions 10 to 22, with complete elimination of the “monomeric” peaks. This elution profile is consistent with Nipah V-dependent formation of a multiprotein entity with the size range of ∼300 to 500 kDa (Fig. 2E). Together, these results suggest that Nipah virus V protein-dependent IFN evasion is the result of efficient STAT protein binding and not STAT protein degradation.
Nipah virus V protein alters STAT subcellular localization.
Indirect immunofluorescence was used to examine the subcellular distribution of the Nipah V protein and its induced STAT-containing complexes. Initial examination of the steady- state Nipah V protein distribution revealed a pattern distinct from that previously reported for the SV5 and HPIV2 V proteins (22, 30). Cells were transfected with HA epitope-tagged Nipah virus V protein vector, fixed, permeabilized, and stained with tag-specific antiserum. Instead of nuclear accumulation, the Nipah V protein distribution was found to be almost exclusively cytoplasmic (Fig. 3 and 4).
FIG. 3.
Nipah virus V protein prevents nuclear import of STAT1 and STAT2. (A) 2FTGH cells were transfected with empty vector or HA-tagged Nipah V expression plasmid (+NV) and were either unstimulated (UNT) or treated for 30 min with 1,000 U of IFN-α per ml (+IFNα). Cells were fixed, permeabilized, and stained sequentially for HA and then for STAT1 or STAT2. (B) Independent block on STAT1 translocation. Same as in panel A, but the cells were treated for 30 min with 5 ng of IFN-γ per ml prior to processing for NV (HA), STAT1, and STAT2 fluorescence. Left panels indicate the parental cell line 2FTGH (WT). Right panels indicate the STAT2-deficient daughter cell line, U6A (STAT2−). Arrows indicate Nipah V protein-expressing cells.
FIG. 4.
Nipah virus V-STAT complexes accumulate in the cytoplasm by a Crm1 nuclear export system. (A) Localization of V proteins. 2FTGH cells were transfected with the HA-tagged V expression vectors and treated 24 h later with LMB (10 ng/ml) for 3 (+) or left untreated (−), fixed, permeabilized, and stained as described in the legend to Fig. 3. (B) Localization of STAT proteins and Nipah virus V in the presence of LMB. Cells were processed as described above, but double staining was used to visualize effects on STAT1 and STAT2 proteins. (C) Nipah virus V blocks IFN-α signaling in the presence of LMB. 2FTGH cells were transfected with an ISRE-luciferase reporter gene, empty vector, SV5 V (SV), or Nipah virus V (NV) expression vectors. The cells were treated with 1,000 U of IFN-α per ml and/or 10 ng of LMB per ml (+) or left untreated (−) prior to lysis and luciferase assays, as in Fig. 1.
The cytoplasmic steady-state accumulation pattern and strong STAT binding by Nipah virus V protein suggested a hypothesis that Nipah virus V protein might influence the localization of the STAT proteins. Indirect immunofluorescence was carried out to directly examine the effect of Nipah virus V protein expression on IFN-induced STAT1 and STAT2 nuclear translocation. The IFN-α/β-activated ISGF3 transcription complex consists of a heterooligomer containing tyrosine-phosphorylated STAT1 and STAT2. STAT2 is cytoplasmically localized prior to IFN treatment and rapidly redistributes to the nucleus following IFN stimulation (23) (Fig. 3A). The distribution of STAT1 is both cytoplasmic and nuclear in unstimulated cells, a result of signal-independent basal nuclear shuttling. However, STAT1 efficiently redistributes to the nucleus following IFN stimulation (17) (Fig. 3A).
The effect of Nipah virus V protein on STAT1 distribution in human 2fTGH cells was immediately apparent in unstimulated cells, since the basal level of nuclear STAT1 protein was absent in cells expressing Nipah virus V protein, producing a more complete STAT1 cytoplasmic localization pattern (Fig. 3A). Treatment of cells with IFN-α resulted in a rapid nuclear translocation of both STAT1 and STAT2. Expression of Nipah virus V protein in the cell completely prevented the STAT nuclear accumulation in response to IFN-α, resulting in a cytoplasmic distribution of both STAT1 and STAT2. This result provides a plausible mechanistic basis for the loss of IFN signaling responses in Nipah virus V protein-expressing cells.
In contrast to the activation of the heterotrimeric ISGF3 complex by IFN-α/β, STAT1 homodimers are the active complex formed in response to IFN-γ, and this STAT1 homodimer rapidly translocates to the nucleus to activate IFN-γ-responsive genes. Expression of Nipah virus V protein also efficiently prevented nuclear accumulation of STAT1 following IFN-γ stimulation (Fig. 3B), consistent with the observed suppression of IFN-γ signaling (Fig. 1C). This finding indicates that the mechanistic basis for Nipah virus V protein suppression of IFN-γ signaling is the result of prohibiting STAT1 protein nuclear transport.
Available evidence indicates that V protein-dependent IFN antagonism and STAT degradation induced by SV5 or HPIV2 infection relies on a multiprotein complex that requires both STAT1 and STAT2 for either virus to degrade its individual STAT target. For example, SV5 cannot degrade STAT1 in cells lacking STAT2 and HPIV2 cannot degrade STAT2 in cells lacking STAT1 (21). To test whether Nipah virus V protein actions against STAT1 are similarly contingent on the availability of cellular STAT2 protein, the IFN-γ-activated distribution of STAT1 was tested in the absence of STAT2. In STAT2-deficient U6A cells, activated STAT1 still accumulated in the nucleus in response to IFN-γ, but expression of Nipah virus V protein abolished IFN-γ-dependent STAT1 nuclear translocation (Fig. 3B). Therefore, Nipah virus V protein is capable of suppressing IFN-γ signaling through STAT1 independent of cellular STAT2 and ISGF3 signals.
Distribution of Nipah V and STATs is regulated by an LMB-sensitive nuclear export system.
The mechanistic basis of Nipah virus V protein subcellular distribution and STAT redistribution was further explored by indirect immunofluorescence (Fig. 4A, top panels). Cells transfected with expression vectors for epitope-tagged V proteins from Nipah virus, SV5, and HPIV2 were fixed, permeabilized, and stained with tag-specific antiserum. HPIV2 and SV5 V proteins both exhibited characteristic nuclear and cytoplasmic localization patterns, as described previously (22, 30). Strikingly, Nipah virus V protein was detected exclusively in the cytoplasm. Treatment of cells with an inhibitor of Crm1-dependent nuclear export, LMB, resulted in redistribution of the Nipah virus V protein to the nucleus, demonstrating that the steady-state Nipah virus V protein cytoplasmic accumulation was the result of nuclear- cytoplasmic shuttling with net export. The distribution of HPIV2 and SV5 V proteins was not altered by LMB treatment (Fig. 4A, bottom panels). The subcellular distribution of Nipah virus V protein differs significantly from that of the other paramyxovirus V proteins, consistent with its distinct mechanisms of action in the host cell.
The effect of LMB-induced Nipah virus V protein redistribution on steady-state localization of STAT proteins was next tested. Treatment of cells with LMB results in a distinct increase in the steady-state nuclear accumulation of both STAT1 and STAT2, in agreement with prior studies of STAT1 protein trafficking (2, 16, 18). The signals controlling STAT2 distribution in the cell have not be previously described, but the results implicate a Crm1-dependent pathway in STAT2 cytoplasmic accumulation (Fig. 4B). The expression of Nipah virus V protein did not superimpose any apparent restriction of STAT protein redistribution in the LMB-treated cells, since treatment of cells with LMB allowed both STAT1 and STAT2 to accumulate in the nucleus, regardless of Nipah V protein expression.
To determine the effect of LMB-mediated STAT reorganization on the IFN antagonist activity of Nipah virus V protein, the nuclear export inhibitor was used in the context of an IFN- dependent reporter gene assay. LMB-induced redistribution had no effect on the ability of Nipah virus V to block ISRE-dependent transcription in the reporter gene assay (Fig. 4C). However, it is noteworthy that LMB treatment did have the unexpected effect of partially disrupting SV5 V protein-mediated IFN antagonism. Together, these findings indicate that a primary action of Nipah virus V protein is to convert STAT proteins to high-molecular-weight complexes, and these complexes normally accumulate in the cytoplasm of Nipah V-expressing cells. We infer from the continued antagonistic action despite LMB-mediated nuclear accumulation that these complexes might also shuttle between the cytoplasm and the nucleus.
Nipah virus V protein prevents STAT activation.
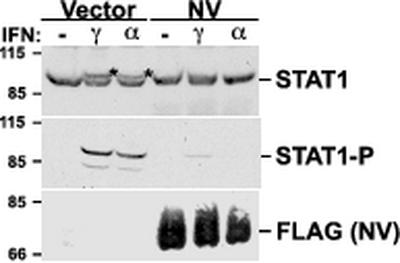
One consequence of Nipah virus V-induced STAT protein complex formation could be the sequestration of STAT proteins, preventing participation in their normal IFN-induced signaling pathways. To directly test for effects of Nipah V protein expression on STAT1 protein activation, IFN-induced STAT1 protein tyrosine phosphorylation was analyzed by immunoblotting. Control 293T cells or cells expressing Nipah virus V protein were left unstimulated or treated with IFN-α or IFN-γ for 30 min prior to lysate preparation. STAT1 activation by IFN-α and IFN-γ was observed in the total STAT1 immunoblot of the cell extracts, evidenced by the IFN-induced appearance of a slower-migrating STAT1 species previously identified as activated STAT1 (25, 26) (Fig. 5). More specifically, this slower-migrating STAT1 is associated with phosphorylation of STAT1 tyrosine residue 701, as demonstrated by probing with a modification-specific phosphopeptide antiserum (Fig. 5, middle panel). In the cells expressing Nipah virus V protein, the level of tyrosine 701-phosphorylated STAT1 protein was greatly reduced, with the minor detected phosphorylation most probably arising from the low percentage of untransfected cells on the plate. This result clearly demonstrates that the Nipah virus V protein expression interferes with the activation of STAT1 signaling, by capture and sequestration of the STATs in the high-molecular-weight complexes.
FIG. 5.
Nipah virus V protein inhibits activating tyrosine phosphorylation of STAT1. Individual 60-mm plates of 293T cells were transfected with 15 μg of empty vector or FLAG-tagged Nipah virus V expression vector (NV). Cells were treated 24 h later with 1,000 U of IFN-α per ml or 5 ng of IFN-γ per ml for 30 min prior to lysis. Lysates (50 μg of total protein) were separated by SDS-PAGE (7% polyacrylamide), transferred to nitrocellulose, and visualized by immunoblotting with antisera to STAT1, phosphotyrosine 701-modified STAT1 (STAT1-P), and FLAG epitope tag.
DISCUSSION
Animal viruses employ a broad range of anti-immune strategies to enable efficient replication, and rapid identification of the host evasion mechanisms exploited by newly emerged pathogens is essential for the timely development of treatment strategies. The transfer of Nipah virus from isolated animal reservoirs into the human population provides an excellent model for studying host-pathogen interactions in emerging infectious agents. Expression of Nipah virus V protein from a cDNA clone reveals that the protein is distinct from other paramyxovirus V proteins. We found that Nipah V protein accumulates in the cytoplasm of cells by an active transport mechanism involving the Crm1 nuclear export system. This cytoplasmic distribution is not observed for the STAT-targeting V proteins encoded by SV5 or HPIV2, which accumulate in the cell nucleus. Nonetheless, the difference in subcellular distribution does not correlate with a difference in cellular function. The results clearly demonstrate that the V protein of Nipah virus is an efficient inhibitor of both IFN-α and IFN-γ signal transduction, rendering cells insensitive to stimulation with either cytokine. Mechanistically, however, Nipah virus V protein has a unique anti-STAT activity not observed for the other paramyxoviruses. Nipah virus V protein does not target STAT1 or STAT2 for proteasomal degradation. Instead, the Nipah virus V protein efficiently binds both STATs, converting them into high-molecular-weight complexes.
The observed IFN inhibition is a consequence of efficient sequestration of the STAT proteins in the cytoplasm. The Nipah V-STAT complexes are inhibitory for STAT protein activation at the receptor-kinase complex, as evidenced by the lack of site-specific STAT1 tyrosine 701 phosphorylation in cells expressing Nipah virus V protein. The finding that Nipah V remains an effective IFN antagonist despite nuclear redistribution by LMB suggests that these complexes are free to move between the nucleus and cytoplasm, with an export rate that is greater than the import rate. Enabling the STATs to accumulate in the nucleus by LMB treatment is not sufficient to reverse the Nipah V-induced IFN antagonism, suggesting that the protein interactions that determine complex formation are maintained in the LMB-treated cells. These data suggest that the complex itself is the critical feature resulting in nuclear exclusion through alteration of the rates of STAT protein flux through the nuclear pore complex.
The estimated size of Nipah V-induced STAT complex(es) is in the range of approximately 300 to 500 kDa. While the exact nature and composition of these complexes cannot be revealed by this initial experiment, the size is consistent with at least a trimeric Nipah virus V protein-STAT1-STAT2 complex. The finding that STAT1 is antagonized independently of STAT2 may indicate that separate Nipah V-STAT1 and Nipah V-STAT2 complexes exist in the cytoplasm, providing for the possibility that these complexes include additional cellular components. Regardless of the precise molecular composition of these Nipah V-induced complexes, the results for IFN signaling are profound. Nipah V expression prevents STAT1 activation by tyrosine phosphorylation and blocks nuclear translocation of both STAT1 and STAT2.
The effects of Nipah virus V protein on STAT1 and STAT2 activation and subcellular distribution described here have been observed only in cells transiently expressing Nipah V protein outside the context of a Nipah virus infection. It remains to be formally demonstrated that the Nipah V protein acts similarly in the context of Nipah virus infection, and it is possible that in infected cells the V protein achieves a different subcellular distribution or carries out additional anticellular or unrelated activities in concert with additional expressed viral proteins. It is noteworthy that for other paramyxoviruses, the V protein anti-STAT activities assayed in infected cells match closely to assays of the V protein functions when expressed in isolation. Therefore, the observed Nipah V IFN antagonism mechanisms probably reflect the actual activity of this protein during virus infection.
Despite the large number of strategies evolved by pathogens to avoid host immune responses, blocking cytokine signal transduction through STAT protein cytoplasmic complex formation has not been observed in other viruses. Acquisition of this unique mechanism for IFN evasion that can subdue STAT1- and STAT2-dependent IFN-α and IFN-γ signals may be one reason accounting for the emergence of Nipah virus as a fatal human pathogen. Since the ability to interfere with host defense response mechanisms is essential for virus pathogenesis, these findings further suggest that the V protein interaction with STATs might be a valuable target for pharmaceutical intervention in emerging paramyxovirus diseases and that ablation of the Nipah virus V gene using a reverse genetic system might result in attenuated Nipah virus that could be used for vaccine production.
Acknowledgments
We are grateful to Paul Rota for providing Nipah virus V cDNA, to George Stark for providing cell lines, to Silviu Tamasdan for assisting with gel filtration, and to Ravi Iyengar, Bob Lamb, Adrian Ting, Horvath laboratory members, and anonymous reviewers for comments on the manuscript and helpful discussions.
This work was supported by research grants to C.M.H. from the American Cancer Society (Research Scholar grant GMC-103079) and NIH grants AI-48722 and AI-50707. J.J.R. is supported by the Integrated Training Program in Pharmacological Sciences (GM-62754). Confocal microscopy at Mount Sinai is partially supported by NIH Shared Instrumentation grant 1 S10 RR0 9145 and NSF Major Research Instrumentation grant DBI-9724504.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 4.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 5.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 6.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, B. T. (ed.). 2001. Introduction to current focus on Hendra and Nipah viruses. Microbes Infect. 3:277-278. [DOI] [PubMed]
- 9.Fu, X.-Y., D. S. Kessler, S. A. Veals, D. A. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon-alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 87:8555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano, M., M. Kaito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 11.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal Cys-rich region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 1177-1204. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 13.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 15:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 16.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, T., A. Begitt, I. Lodige, M. van Rossum, and U. Vinkemeier. 2002. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mowen, K., and M. David. 2000. Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol. 20:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parisien, J.-P., J. F. Lau, and C. M. Horvath. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed]
- 20.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 21.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2, but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 23.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809-813. [DOI] [PubMed] [Google Scholar]
- 24.Sehgal, P. B. 2000. STAT-signalling through the cytoplasmic compartment: consideration of a new paradigm. Cell Signal. 12:525-535. [DOI] [PubMed] [Google Scholar]
- 25.Shuai, K., C. M. Horvath, L. H. Tsai-Huang, S. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 26.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 27.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 28.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 29.Wang, L. F., M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski, and B. T. Eaton. 2000. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74:9972-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, N., M. Kawano, M. Tsurudome, S. Kusagawa, M. Nishio, H. Komada, T. Shima, and Y. Ito. 1996. Identification of the sequences responsible for nuclear targeting of the V protein of human parainfluenza virus type 2. J. Gen. Virol. 77:327-338. [DOI] [PubMed] [Google Scholar]